Genetic and epigenetic regulation of the organic cation transporter 3, SLC22A3
- PMID: 22231567
- PMCID: PMC3396779
- DOI: 10.1038/tpj.2011.60
Genetic and epigenetic regulation of the organic cation transporter 3, SLC22A3
Abstract
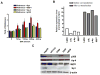
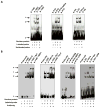
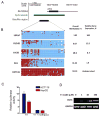
Human organic cation transporter 3 (OCT3 and SLC22A3) mediates the uptake of many important endogenous amines and basic drugs in a variety of tissues. OCT3 is identified as one of the important risk loci for prostate cancer, and is markedly underexpressed in aggressive prostate cancers. The goal of this study was to identify genetic and epigenetic factors in the promoter region that influence the expression level of OCT3. Haplotypes that contained the common variants, g.-81G>delGA (rs60515630) (minor allele frequency 11.5% in African American) and g.-2G>A (rs555754) (minor allele frequency>30% in all ethnic groups) showed significant increases in luciferase reporter activities and exhibited stronger transcription factor-binding affinity than the haplotypes that contained the major alleles. Consistent with the reporter assays, OCT3 messenger RNA expression levels were significantly higher in Asian (P<0.001) and Caucasian (P<0.05) liver samples from individuals who were homozygous for g.-2A/A in comparison with those homozygous for the g.-2G/G allele. Studies revealed that the methylation level in the basal promoter region of OCT3 was associated with OCT3 expression level and tumorigenesis capability in various prostate cancer cell lines. The methylation level of the OCT3 promoter was higher in 62% of prostate tumor samples compared with matched normal samples. Our studies demonstrate that genetic polymorphisms in the proximal promoter region of OCT3 alter the transcription rate of the gene and may be associated with altered expression levels of OCT3 in human liver. Aberrant methylation contributes to the reduced expression of OCT3 in prostate cancer.
Conflict of interest statement
Authors declare there are no competing financial interests in relation to the work.
Figures
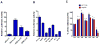


References
-
- Tregouet DA, Konig IR, Erdmann J, Munteanu A, Braund PS, Hall AS, et al. Genome-wide haplotype association study identifies the SLC22A3-LPAL2-LPA gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–5. - PubMed
-
- Lazar A, Walitza S, Jetter A, Gerlach M, Warnke A, Herpertz-Dahlmann B, et al. Novel mutations of the extraneuronal monoamine transporter gene in children and adolescents with obsessive-compulsive disorder. Int J Neuropsychopharmacol. 2008;11:35–48. - PubMed
-
- Eeles RA, Kote-Jarai Z, Giles GG, Al Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nature Genet. 2008;40:316–21. - PubMed
-
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

