Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice
- PMID: 22232209
- PMCID: PMC3266788
- DOI: 10.1172/JCI59227
Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice
Abstract
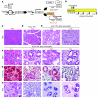
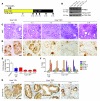
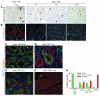
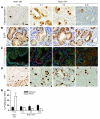
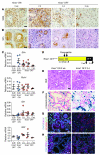
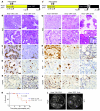
Pancreatic cancer is almost invariably associated with mutations in the KRAS gene, most commonly KRASG12D, that result in a dominant-active form of the KRAS GTPase. However, how KRAS mutations promote pancreatic carcinogenesis is not fully understood, and whether oncogenic KRAS is required for the maintenance of pancreatic cancer has not been established. To address these questions, we generated two mouse models of pancreatic tumorigenesis: mice transgenic for inducible KrasG12D, which allows for inducible, pancreas-specific, and reversible expression of the oncogenic KrasG12D, with or without inactivation of one allele of the tumor suppressor gene p53. Here, we report that, early in tumorigenesis, induction of oncogenic KrasG12D reversibly altered normal epithelial differentiation following tissue damage, leading to precancerous lesions. Inactivation of KrasG12D in established precursor lesions and during progression to cancer led to regression of the lesions, indicating that KrasG12D was required for tumor cell survival. Strikingly, during all stages of carcinogenesis, KrasG12D upregulated Hedgehog signaling, inflammatory pathways, and several pathways known to mediate paracrine interactions between epithelial cells and their surrounding microenvironment, thus promoting formation and maintenance of the fibroinflammatory stroma that plays a pivotal role in pancreatic cancer. Our data establish that epithelial KrasG12D influences multiple cell types to drive pancreatic tumorigenesis and is essential for tumor maintenance. They also strongly support the notion that inhibiting KrasG12D, or its downstream effectors, could provide a new approach for the treatment of pancreatic cancer.
Figures

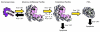
References
-
- National Cancer Institute. Surveillance, Epidemiology and End Results (SEER) Program web site. http://seer.cancer.gov . Accessed December 7, 2011.
-
- American Cancer Society. Cancer Facts and Figures website. http://www.cancer.org/Research/CancerFactsFigures . Accessed December 7, 2011.
-
- Hishinuma S, Ogata Y, Tomikawa M, Ozawa I, Hirabayashi K, Igarashi S. Patterns of recurrence after curative resection of pancreatic cancer, based on autopsy findings. J Gastrointest Surg. 2006;10(4):511–518. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

