Protein docking prediction using predicted protein-protein interface
- PMID: 22233443
- PMCID: PMC3287255
- DOI: 10.1186/1471-2105-13-7
Protein docking prediction using predicted protein-protein interface
Abstract
Background: Many important cellular processes are carried out by protein complexes. To provide physical pictures of interacting proteins, many computational protein-protein prediction methods have been developed in the past. However, it is still difficult to identify the correct docking complex structure within top ranks among alternative conformations.
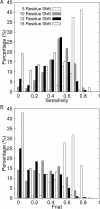
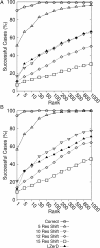
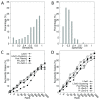
Results: We present a novel protein docking algorithm that utilizes imperfect protein-protein binding interface prediction for guiding protein docking. Since the accuracy of protein binding site prediction varies depending on cases, the challenge is to develop a method which does not deteriorate but improves docking results by using a binding site prediction which may not be 100% accurate. The algorithm, named PI-LZerD (using Predicted Interface with Local 3D Zernike descriptor-based Docking algorithm), is based on a pair wise protein docking prediction algorithm, LZerD, which we have developed earlier. PI-LZerD starts from performing docking prediction using the provided protein-protein binding interface prediction as constraints, which is followed by the second round of docking with updated docking interface information to further improve docking conformation. Benchmark results on bound and unbound cases show that PI-LZerD consistently improves the docking prediction accuracy as compared with docking without using binding site prediction or using the binding site prediction as post-filtering.
Conclusion: We have developed PI-LZerD, a pairwise docking algorithm, which uses imperfect protein-protein binding interface prediction to improve docking accuracy. PI-LZerD consistently showed better prediction accuracy over alternative methods in the series of benchmark experiments including docking using actual docking interface site predictions as well as unbound docking cases.
Figures




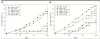

References
-
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. - DOI - PubMed
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

