A searchable cross-platform gene expression database reveals connections between drug treatments and disease
- PMID: 22233519
- PMCID: PMC3305579
- DOI: 10.1186/1471-2164-13-12
A searchable cross-platform gene expression database reveals connections between drug treatments and disease
Abstract
Background: Transcriptional data covering multiple platforms and species is collected and processed into a searchable platform independent expression database (SPIED). SPIED consists of over 100,000 expression fold profiles defined independently of control/treatment assignment and mapped to non-redundant gene lists. The database is thus searchable with query profiles defined over genes alone. The motivation behind SPIED is that transcriptional profiles can be quantitatively compared and ranked and thus serve as effective surrogates for comparing the underlying biological states across multiple experiments.
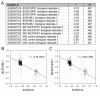
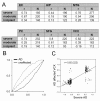
Results: Drug perturbation, cancer and neurodegenerative disease derived transcriptional profiles are shown to be effective descriptors of the underlying biology as they return related drugs and pathologies from SPIED. In the case of Alzheimer's disease there is high transcriptional overlap with other neurodegenerative conditions and rodent models of neurodegeneration and nerve injury. Combining the query signature with correlating profiles allows for the definition of a tight neurodegeneration signature that successfully highlights many neuroprotective drugs in the Broad connectivity map.
Conclusions: Quantitative querying of expression data from across the totality of deposited experiments is an effective way of discovering connections between different biological systems and in particular that between drug action and biological disease state. Examples in cancer and neurodegenerative conditions validate the utility of SPIED.
Figures


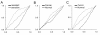
References
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al.Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

