Preferential nucleosome occupancy at high values of DNA helical rise
- PMID: 22233711
- PMCID: PMC3276262
- DOI: 10.1093/dnares/dsr043
Preferential nucleosome occupancy at high values of DNA helical rise
Abstract
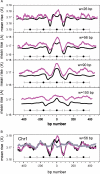
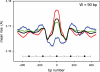
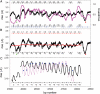
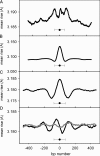
Nucleosomes are the basic structural units of eukaryotic chromatin and play a key role in the regulation of gene expression. Nucleosome formation depends on several factors, including properties of the sequence itself, but also physical constraints and epigenetic factors such as chromatin-remodelling enzymes. In this view, a sequence-dependent approach is able to capture a general tendency of a region to bind a histone octamer. A reference data set of positioned nucleosomes of Saccharomyces cerevisiae was used to study the role of DNA helical rise in histone-DNA interaction. Genomic sequences were transformed into arrays of helical rise values by a tetranucleotide code and then turned into profiles of mean helical rise values. These profiles resemble maps of nucleosome occupancy, suggesting that intrinsic histone-DNA interactions are linked to helical rise. The obtained results show that preferential nucleosome occupancy occurs where the mean helical rise reaches its largest values. Mean helical rise profiles obtained by using maps of positioned nucleosomes of the Drosophila melanogaster and Plasmodium falciparum genomes, as well as Homo sapiens chromosome 20 confirm that nucleosomes are mainly located where the mean helical rise reaches its largest values.
Figures
Similar articles
-
DNA physical properties determine nucleosome occupancy from yeast to fly.Nucleic Acids Res. 2008 Jun;36(11):3746-56. doi: 10.1093/nar/gkn262. Epub 2008 May 17. Nucleic Acids Res. 2008. PMID: 18487627 Free PMC article.
-
Prediction of nucleosome DNA formation potential and nucleosome positioning using increment of diversity combined with quadratic discriminant analysis.Chromosome Res. 2010 Nov;18(7):777-85. doi: 10.1007/s10577-010-9160-9. Epub 2010 Oct 16. Chromosome Res. 2010. PMID: 20953693
-
In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae.PLoS Genet. 2012;8(6):e1002771. doi: 10.1371/journal.pgen.1002771. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737086 Free PMC article.
-
Nucleosome Positioning and Spacing: From Mechanism to Function.J Mol Biol. 2021 Mar 19;433(6):166847. doi: 10.1016/j.jmb.2021.166847. Epub 2021 Feb 2. J Mol Biol. 2021. PMID: 33539878 Review.
-
Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution.Biochem Soc Trans. 2012 Apr;40(2):377-82. doi: 10.1042/BST20110730. Biochem Soc Trans. 2012. PMID: 22435815 Review.
Cited by
-
A survey of k-mer methods and applications in bioinformatics.Comput Struct Biotechnol J. 2024 May 21;23:2289-2303. doi: 10.1016/j.csbj.2024.05.025. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38840832 Free PMC article. Review.
-
Atomistic Insights Into Interaction of Doxorubicin With DNA: From Duplex to Nucleosome.J Comput Chem. 2025 Jan 30;46(3):e70035. doi: 10.1002/jcc.70035. J Comput Chem. 2025. PMID: 39865531 Free PMC article.
-
Nullomers and High Order Nullomers in Genomic Sequences.PLoS One. 2016 Dec 1;11(12):e0164540. doi: 10.1371/journal.pone.0164540. eCollection 2016. PLoS One. 2016. PMID: 27906971 Free PMC article.
References
-
- Lohr D., Fatchell K., Van-Holde K.E. On the occurrence of nucleosome phasing in chromatin. Cell. 1977;12:829–36. - PubMed
-
- Trifonov E.N. Structure of DNA in chromatin. Eur. J. Cell Biol. 1980;22:74.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

