Cloning and heterologous expression of Plasmodium ovale dihydrofolate reductase-thymidylate synthase gene
- PMID: 22234170
- PMCID: PMC3444756
- DOI: 10.1016/j.parint.2011.12.004
Cloning and heterologous expression of Plasmodium ovale dihydrofolate reductase-thymidylate synthase gene
Abstract
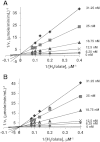
Plasmodial bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS) is a validated antimalarial drug target. In this study, expression of the putative dhfr-ts of Plasmodium ovale rescued the DHFR chemical knockout and a TS null bacterial strain, demonstrating its DHFR and TS catalytic functions. PoDHFR-TS was expressed in Escherichia coli BL21 (DE3) and affinity purified by Methotrexate Sepharose column. Biochemical and enzyme kinetics characterizations indicated that PoDHFR-TS is similar to other plasmodial enzymes, albeit with lower catalytic activity but better tolerance of acidic pH. Importantly, the PoDHFR from Thai isolate EU266602 remains sensitive to the antimalarials pyrimethamine and cycloguanil, in contrast to P. falciparum and P. vivax isolates where resistance to these drugs is widespread.
Copyright © 2011 Elsevier Ireland Ltd. All rights reserved.
Figures

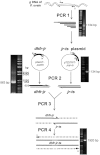


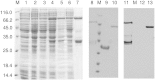
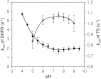
References
-
- Al-Maktari M.T., Bassiouny H.K., Al-Hamd Z.S., Assabri A.M., El-Massry A.G., Shatat H.Z. Malaria status in Al-Hodeidah Governorate, Yemen: malariometric parasitic survey & chloroquine resistance P. falciparum local strain. Journal of the Egyptian Society of Parasitology. 2003;33:361–372. - PubMed
-
- Win T.T., Lin K., Mizuno S., Zhou M., Liu Q., Ferreira M.U. Wide distribution of Plasmodium ovale in Myanmar. Tropical Medicine & International Health. 2002;7:231–239. - PubMed
-
- Galinski M.R., Barnwell J.W. Monkey malaria kills four humans. Trends in Parasitology. 2009;25:200–204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

