Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype
- PMID: 22234187
- PMCID: PMC3280564
- DOI: 10.1038/emboj.2011.485
Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype
Abstract
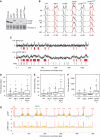
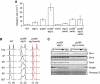
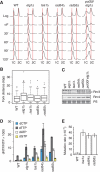
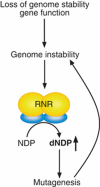
The integrity of the genome depends on diverse pathways that regulate DNA metabolism. Defects in these pathways result in genome instability, a hallmark of cancer. Deletion of ELG1 in budding yeast, when combined with hypomorphic alleles of PCNA results in spontaneous DNA damage during S phase that elicits upregulation of ribonucleotide reductase (RNR) activity. Increased RNR activity leads to a dramatic expansion of deoxyribonucleotide (dNTP) pools in G1 that allows cells to synthesize significant fractions of the genome in the presence of hydroxyurea in the subsequent S phase. Consistent with the recognized correlation between dNTP levels and spontaneous mutation, compromising ELG1 and PCNA results in a significant increase in mutation rates. Deletion of distinct genome stability genes RAD54, RAD55, and TSA1 also results in increased dNTP levels and mutagenesis, suggesting that this is a general phenomenon. Together, our data point to a vicious circle in which mutations in gatekeeper genes give rise to genomic instability during S phase, inducing expansion of the dNTP pool, which in turn results in high levels of spontaneous mutagenesis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 - PubMed
-
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

