p38 MAPK inhibition suppresses the TLR-hypersensitive phenotype in FANCC- and FANCA-deficient mononuclear phagocytes
- PMID: 22234699
- PMCID: PMC3311243
- DOI: 10.1182/blood-2011-06-354647
p38 MAPK inhibition suppresses the TLR-hypersensitive phenotype in FANCC- and FANCA-deficient mononuclear phagocytes
Abstract
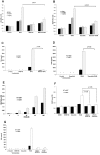
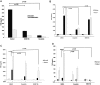
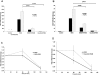
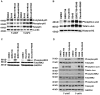
Fanconi anemia, complementation group C (FANCC)-deficient hematopoietic stem and progenitor cells are hypersensitive to a variety of inhibitory cytokines, one of which, TNFα, can induce BM failure and clonal evolution in Fancc-deficient mice. FANCC-deficient macrophages are also hypersensitive to TLR activation and produce TNFα in an unrestrained fashion. Reasoning that suppression of inhibitory cytokine production might enhance hematopoiesis, we screened small molecules using TLR agonist-stimulated FANCC- and Fanconi anemia, complementation group A (FANCA)-deficient macrophages containing an NF-κB/AP-1-responsive reporter gene (SEAP). Of the 75 small molecules screened, the p38 MAPK inhibitor BIRB 796 and dasatinib potently suppressed TLR8-dependent expression of the reporter gene. Fanconi anemia (FA) macrophages were hypersensitive to the TLR7/8 activator R848, overproducing SEAP and TNFα in response to all doses of the agonist. Low doses (50nM) of both agents inhibited p38 MAPK-dependent activation of MAPKAPK2 (MK2) and suppressed MK2-dependent TNFα production without substantially influencing TNFα gene transcription. Overproduction of TNFα by primary FA cells was likewise suppressed by these agents and involved inhibition of MK2 activation. Because MK2 is also known to influence production and/or sensitivity to 2 other suppressive factors (MIP-1α and IFNγ) to which FA hematopoietic progenitor cells are uniquely vulnerable, targeting of p38 MAPK in FA hematopoietic cells is a rational objective for preclinical evaluation.
Figures

References
-
- de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668(1-2):11–19. - PubMed
-
- Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43(2):138–141. - PubMed
-
- Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

