A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in Candida glabrata
- PMID: 22234857
- PMCID: PMC3316643
- DOI: 10.1534/genetics.111.138099
A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in Candida glabrata
Abstract
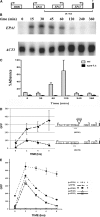
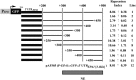
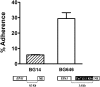
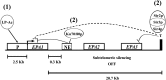
Candida glabrata, an opportunistic fungal pathogen, adheres to mammalian epithelial cells; adherence is mediated primarily by the Epa1 adhesin. EPA1 is a member of a large gene family of ≈ 23 paralogues, which encode putative adhesins. In this study, we address how EPA1 transcription is regulated. Our data show that EPA1 expression is subject to two distinct negative regulatory mechanisms. EPA1 transcription is repressed by subtelomeric silencing: the Sir complex (Sir2-Sir4), Rap1, Rif1, yKu70, and yKu80 are required for full repression. Activation of EPA1 occurs immediately after dilution of stationary phase (SP) cells into fresh media; however, transcription is rapidly repressed again, limiting expression to lag phase, just as the cells exit stationary phase. This repression following lag phase requires a cis-acting regulatory negative element (NE) located in the EPA1 3'-intergenic region and is independent of telomere proximity. Bioinformatic analysis shows that there are 10 copies of the NE-like sequence in the C. glabrata genome associated with other EPA genes as well as non-EPA genes.
Figures


Similar articles
-
Abf1 negatively regulates the expression of EPA1 and affects adhesion in Candida glabrata.J Med Microbiol. 2024 Oct;73(10). doi: 10.1099/jmm.0.001905. J Med Microbiol. 2024. PMID: 39360930
-
Chromatin Loop Formation Induced by a Subtelomeric Protosilencer Represses EPA Genes in Candida glabrata.Genetics. 2018 Sep;210(1):113-128. doi: 10.1534/genetics.118.301202. Epub 2018 Jul 12. Genetics. 2018. PMID: 30002080 Free PMC article.
-
yKu70/yKu80 and Rif1 regulate silencing differentially at telomeres in Candida glabrata.Eukaryot Cell. 2008 Dec;7(12):2168-78. doi: 10.1128/EC.00228-08. Epub 2008 Oct 3. Eukaryot Cell. 2008. PMID: 18836091 Free PMC article.
-
Local and regional chromatin silencing in Candida glabrata: consequences for adhesion and the response to stress.FEMS Yeast Res. 2015 Sep;15(6):fov056. doi: 10.1093/femsyr/fov056. Epub 2015 Jun 29. FEMS Yeast Res. 2015. PMID: 26122277 Review.
-
The mating type-like loci of Candida glabrata.Rev Iberoam Micol. 2014 Jan-Mar;31(1):30-4. doi: 10.1016/j.riam.2013.09.016. Epub 2013 Nov 16. Rev Iberoam Micol. 2014. PMID: 24252826 Review.
Cited by
-
Structural Hot Spots Determine Functional Diversity of the Candida glabrata Epithelial Adhesin Family.J Biol Chem. 2015 Aug 7;290(32):19597-613. doi: 10.1074/jbc.M115.655654. Epub 2015 Jun 23. J Biol Chem. 2015. PMID: 26105055 Free PMC article.
-
Upregulation of the Adhesin Gene EPA1 Mediated by PDR1 in Candida glabrata Leads to Enhanced Host Colonization.mSphere. 2016 Mar 2;1(2):e00065-15. doi: 10.1128/mSphere.00065-15. eCollection 2016 Mar-Apr. mSphere. 2016. PMID: 27303714 Free PMC article.
-
Trypanosoma brucei RAP1 Has Essential Functional Domains That Are Required for Different Protein Interactions.mSphere. 2020 Feb 26;5(1):e00027-20. doi: 10.1128/mSphere.00027-20. mSphere. 2020. PMID: 32102938 Free PMC article.
-
To Know How a Gene Works, We Need to Redefine It First but then, More Importantly, to Let the Cell Itself Decide How to Transcribe and Process Its RNAs.Int J Biol Sci. 2015 Nov 19;11(12):1413-23. doi: 10.7150/ijbs.13436. eCollection 2015. Int J Biol Sci. 2015. PMID: 26681921 Free PMC article. Review.
-
Telomeric and Sub-Telomeric Structure and Implications in Fungal Opportunistic Pathogens.Microorganisms. 2021 Jun 29;9(7):1405. doi: 10.3390/microorganisms9071405. Microorganisms. 2021. PMID: 34209786 Free PMC article. Review.
References
-
- Ausubel, F., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman et al., 2001 Current Protocols in Molecular Biology. John Wiley & Sons, New York.
-
- Calderwood M. S., Gannoun-Zaki L., Wellems T. E., Deitsch K. W., 2003. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278: 34125–34132 - PubMed
-
- Castaño I., Pan S. J., Zupancic M., Hennequin C., Dujon B., et al. , 2005. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 55: 1246–1258 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

