Gene genealogies within a fixed pedigree, and the robustness of Kingman's coalescent
- PMID: 22234858
- PMCID: PMC3316654
- DOI: 10.1534/genetics.111.135574
Gene genealogies within a fixed pedigree, and the robustness of Kingman's coalescent
Abstract
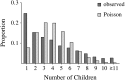
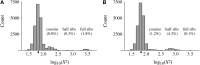
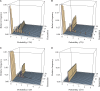
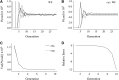
We address a conceptual flaw in the backward-time approach to population genetics called coalescent theory as it is applied to diploid biparental organisms. Specifically, the way random models of reproduction are used in coalescent theory is not justified. Instead, the population pedigree for diploid organisms--that is, the set of all family relationships among members of the population--although unknown, should be treated as a fixed parameter, not as a random quantity. Gene genealogical models should describe the outcome of the percolation of genetic lineages through the population pedigree according to Mendelian inheritance. Using simulated pedigrees, some of which are based on family data from 19th century Sweden, we show that in many cases the (conceptually wrong) standard coalescent model is difficult to reject statistically and in this sense may provide a surprisingly accurate description of gene genealogies on a fixed pedigree. We study the differences between the fixed-pedigree coalescent and the standard coalescent by analysis and simulations. Differences are apparent in recent past, within ≈ <log(2)(N) generations, but then disappear as genetic lineages are traced into the more distant past.
Figures


References
-
- Ball M., Neigel J. E., Avise J. C., 1990. Gene genealogies within organismal pedigrees of random-mating populations. Evolution 44: 360–370 - PubMed
-
- Bittles A. H., Egerbladh I., 2005. The influence of past endogamy and consanguinity on genetic disorders in northern Sweden. Ann. Hum. Genet. 69: 549–558 - PubMed
-
- Cannings C., 1974. The latent roots of certain Markov chains arising in genetics: a new approach. I. Haploid models. Adv. Appl. Probab. 6: 260–290
-
- Cannings C., Thompson E. A., Skolnick M. H., 1978. Probability functions on complex pedigrees. Adv. Appl. Probab. 10: 26–61
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

