An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement
- PMID: 22234889
- PMCID: PMC3317147
- DOI: 10.1101/gr.132233.111
An ancient genomic regulatory block conserved across bilaterians and its dismantling in tetrapods by retrogene replacement
Abstract
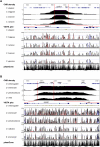
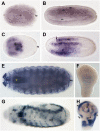
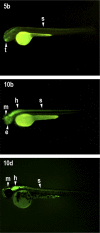
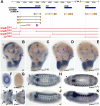
Developmental genes are regulated by complex, distantly located cis-regulatory modules (CRMs), often forming genomic regulatory blocks (GRBs) that are conserved among vertebrates and among insects. We have investigated GRBs associated with Iroquois homeobox genes in 39 metazoans. Despite 600 million years of independent evolution, Iroquois genes are linked to ankyrin-repeat-containing Sowah genes in nearly all studied bilaterians. We show that Iroquois-specific CRMs populate the Sowah locus, suggesting that regulatory constraints underlie the maintenance of the Iroquois-Sowah syntenic block. Surprisingly, tetrapod Sowah orthologs are intronless and not associated with Iroquois; however, teleost and elephant shark data demonstrate that this is a derived feature, and that many Iroquois-CRMs were ancestrally located within Sowah introns. Retroposition, gene, and genome duplication have allowed selective elimination of Sowah exons from the Iroquois regulatory landscape while keeping associated CRMs, resulting in large associated gene deserts. These results highlight the importance of CRMs in imposing constraints to genome architecture, even across large phylogenetic distances, and of gene duplication-mediated genetic redundancy to disentangle these constraints, increasing genomic plasticity.
Figures



References
-
- Abascal F, Zardoya R, Posada D 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 - PubMed
-
- Becker T, Lenhard B 2007. The random versus fragile breakage models of chromosome evolution: a matter of resolution. Mol Genet Genomics 278: 487–491 - PubMed
-
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
