HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter
- PMID: 22234890
- PMCID: PMC3290780
- DOI: 10.1101/gr.128652.111
HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter
Abstract
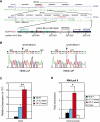
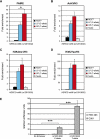
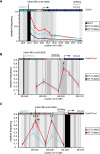
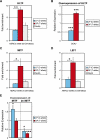
Pigmentation of skin, eye, and hair reflects some of the most evident common phenotypes in humans. Several candidate genes for human pigmentation are identified. The SNP rs12913832 has strong statistical association with human pigmentation. It is located within an intron of the nonpigment gene HERC2, 21 kb upstream of the pigment gene OCA2, and the region surrounding rs12913832 is highly conserved among animal species. However, the exact functional role of HERC2 rs12913832 in human pigmentation is unknown. Here we demonstrate that the HERC2 rs12913832 region functions as an enhancer regulating OCA2 transcription. In darkly pigmented human melanocytes carrying the rs12913832 T-allele, we detected binding of the transcription factors HLTF, LEF1, and MITF to the HERC2 rs12913832 enhancer, and a long-range chromatin loop between this enhancer and the OCA2 promoter that leads to elevated OCA2 expression. In contrast, in lightly pigmented melanocytes carrying the rs12913832 C-allele, chromatin-loop formation, transcription factor recruitment, and OCA2 expression are all reduced. Hence, we demonstrate that allelic variation of a common noncoding SNP located in a distal regulatory element not only disrupts the regulatory potential of this element but also affects its interaction with the relevant promoter. We provide the key mechanistic insight that allele-dependent differences in chromatin-loop formation (i.e., structural differences in the folding of gene loci) result in differences in allelic gene expression that affects common phenotypic traits. This concept is highly relevant for future studies aiming to unveil the functional basis of genetically determined phenotypes, including diseases.
Figures

Comment in
-
Gene expression: Looping together the functions of SNPs.Nat Rev Genet. 2012 Feb 14;13(3):148-9. doi: 10.1038/nrg3180. Nat Rev Genet. 2012. PMID: 22330766 No abstract available.
References
-
- Branicki W, Brudnik U, Wojas-Pelc A 2009. Interactions between HERC2, OCA2 and MC1R may influence human pigmentation phenotype. Ann Hum Genet 73: 160–170 - PubMed
-
- Brilliant MH 2001. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res 14: 86–93 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
