T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription
- PMID: 22235136
- PMCID: PMC3307275
- DOI: 10.1074/jbc.M111.318410
T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription
Abstract
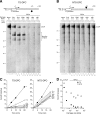
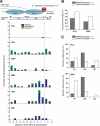
RNA polymerases (RNAPs) transcribe genes through the barrier of nucleoproteins and site-specific DNA-binding proteins on their own or with the aid of accessory factors. Proteins are often covalently trapped on DNA by DNA damaging agents, forming DNA-protein cross-links (DPCs). However, little is known about how immobilized proteins affect transcription. To elucidate the effect of DPCs on transcription, we constructed DNA templates containing site-specific DPCs and performed in vitro transcription reactions using phage T7 RNAP. We show here that DPCs constitute strong but not absolute blocks to in vitro transcription catalyzed by T7 RNAP. More importantly, sequence analysis of transcripts shows that RNAPs roadblocked not only by DPCs but also by the stalled leading RNAP become highly error prone and generate mutations in the upstream intact template regions. This contrasts with the transcriptional mutations induced by conventional DNA lesions, which are delivered to the active site or its proximal position in RNAPs and cause direct misincorporation. Our data also indicate that the trailing RNAP stimulates forward translocation of the stalled leading RNAP, promoting the translesion bypass of DPCs. The present results provide new insights into the transcriptional fidelity and mutual interactions of RNAPs that encounter persistent roadblocks.
Figures


References
-
- Barker S., Weinfeld M., Murray D. (2005) DNA-protein cross-links. Their induction, repair, and biological consequences. Mutat. Res. 589, 111–135 - PubMed
-
- Ide H., Shoulkamy M. I., Nakano T., Miyamoto-Matsubara M., Salem A. M. (2011) Repair and biochemical effects of DNA-protein cross-links. Mutat. Res. 711, 113–122 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

