Analysis of epitopes on dengue virus envelope protein recognized by monoclonal antibodies and polyclonal human sera by a high throughput assay
- PMID: 22235356
- PMCID: PMC3250511
- DOI: 10.1371/journal.pntd.0001447
Analysis of epitopes on dengue virus envelope protein recognized by monoclonal antibodies and polyclonal human sera by a high throughput assay
Abstract
Background: The envelope (E) protein of dengue virus (DENV) is the major target of neutralizing antibodies and vaccine development. While previous studies on domain III or domain I/II alone have reported several epitopes of monoclonal antibodies (mAbs) against DENV E protein, the possibility of interdomain epitopes and the relationship between epitopes and neutralizing potency remain largely unexplored.
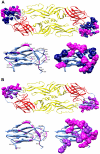
Methodology/principal findings: We developed a dot blot assay by using 67 alanine mutants of predicted surface-exposed E residues as a systematic approach to identify epitopes recognized by mAbs and polyclonal sera, and confirmed our findings using a capture-ELISA assay. Of the 12 mouse mAbs tested, three recognized a novel epitope involving residues (Q211, D215, P217) at the central interface of domain II, and three recognized residues at both domain III and the lateral ridge of domain II, suggesting a more frequent presence of interdomain epitopes than previously appreciated. Compared with mAbs generated by traditional protocols, the potent neutralizing mAbs generated by a new protocol recognized multiple residues in A strand or residues in C strand/CC' loop of DENV2 and DENV1, and multiple residues in BC loop and residues in DE loop, EF loop/F strand or G strand of DENV1. The predominant epitopes of anti-E antibodies in polyclonal sera were found to include both fusion loop and non-fusion residues in the same or adjacent monomer.
Conclusions/significance: Our analyses have implications for epitope-specific diagnostics and epitope-based dengue vaccines. This high throughput method has tremendous application for mapping both intra and interdomain epitopes recognized by human mAbs and polyclonal sera, which would further our understanding of humoral immune responses to DENV at the epitope level.
© 2012 Lin et al.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. - PubMed
-
- World Health Organization. Dengue hemorrhagic fever: Diagnosis, treatment, prevention and control, 3rd ed. Geneva, Switzerland: 2009.
-
- Whitehead SS, Blaney JE, Durbin AP, Murphy BR. Prospects for a dengue virus vaccine. Nat Rev Microbiol. 2007;5:518–528. - PubMed
-
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: the viruses and their replication, In: Knipe DM, Howley PM, editors. Fields virology, 5th ed. Philadelphia: Lippincott William & Wilkins; 2007. pp. 1101–1152.
-
- Heinz FX, Stiasny K. Flavivirus membrane fusion. J Gen Virol. 2006;87:2755–2766. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

