Biomarker robustness reveals the PDGF network as driving disease outcome in ovarian cancer patients in multiple studies
- PMID: 22236809
- PMCID: PMC3298526
- DOI: 10.1186/1752-0509-6-3
Biomarker robustness reveals the PDGF network as driving disease outcome in ovarian cancer patients in multiple studies
Abstract
Background: Ovarian cancer causes more deaths than any other gynecological cancer. Identifying the molecular mechanisms that drive disease progress in ovarian cancer is a critical step in providing therapeutics, improving diagnostics, and affiliating clinical behavior with disease etiology. Identification of molecular interactions that stratify prognosis is key in facilitating a clinical-molecular perspective.
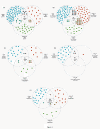
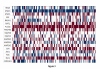
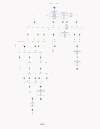
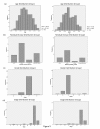
Results: The Cancer Genome Atlas has recently made available the molecular characteristics of more than 500 patients. We used the TCGA multi-analysis study, and two additional datasets and a set of computational algorithms that we developed. The computational algorithms are based on methods that identify network alterations and quantify network behavior through gene expression.We identify a network biomarker that significantly stratifies survival rates in ovarian cancer patients. Interestingly, expression levels of single or sets of genes do not explain the prognostic stratification. The discovered biomarker is composed of the network around the PDGF pathway. The biomarker enables prognosis stratification.
Conclusion: The work presented here demonstrates, through the power of gene-expression networks, the criticality of the PDGF network in driving disease course. In uncovering the specific interactions within the network, that drive the phenotype, we catalyze targeted treatment, facilitate prognosis and offer a novel perspective into hidden disease heterogeneity.
Figures

References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. - PubMed
-
- Ahluwalia A, Yan P, Hurteau JA, Bigsby RM, Jung SH, Huang TH, Nephew KP. DNA methylation and ovarian cancer. I. Analysis of CpG island hypermethylation in human ovarian cancer using differential methylation hybridization. Gynecol Oncol. 2001;82(2):261–268. - PubMed
-
- Bardos A. Treatment of ovarian cancer. 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

