Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance
- PMID: 22236876
- PMCID: PMC3335942
- DOI: 10.4161/cbt.12.12.18671
Glutamine fuels a vicious cycle of autophagy in the tumor stroma and oxidative mitochondrial metabolism in epithelial cancer cells: implications for preventing chemotherapy resistance
Abstract
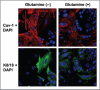
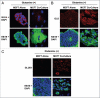
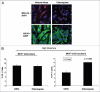
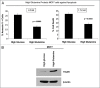
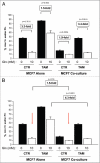
Glutamine metabolism is crucial for cancer cell growth via the generation of intermediate molecules in the tricarboxylic acid (TCA) cycle, antioxidants and ammonia. The goal of the current study was to evaluate the effects of glutamine on metabolism in the breast cancer tumor microenvironment, with a focus on autophagy and cell death in both epithelial and stromal compartments. For this purpose, MCF7 breast cancer cells were cultured alone or co-cultured with non-transformed fibroblasts in media containing high glutamine and low glucose (glutamine +) or under control conditions, with no glutamine and high glucose (glutamine -). Here, we show that MCF7 cells maintained in co-culture with glutamine display increased mitochondrial mass, as compared with control conditions. Importantly, treatment with the autophagy inhibitor chloroquine abolishes the glutamine-induced augmentation of mitochondrial mass. It is known that loss of caveolin-1 (Cav-1) expression in fibroblasts is associated with increased autophagy and an aggressive tumor microenvironment. Here, we show that Cav-1 downregulation which occurs in fibroblasts maintained in co-culture specifically requires glutamine. Interestingly, glutamine increases the expression of autophagy markers in fibroblasts, but decreases expression of autophagy markers in MCF7 cells, indicating that glutamine regulates the autophagy program in a compartment-specific manner. Functionally, glutamine protects MCF7 cells against apoptosis, via the upregulation of the anti-apoptotic and anti-autophagic protein TIGAR. Also, we show that glutamine cooperates with stromal fibroblasts to confer tamoxifen-resistance in MCF7 cancer cells. Finally, we provide evidence that co-culture with fibroblasts (1) promotes glutamine catabolism, and (2) decreases glutamine synthesis in MCF7 cancer cells. Taken together, our findings suggest that autophagic fibroblasts may serve as a key source of energy-rich glutamine to fuel cancer cell mitochondrial activity, driving a vicious cycle of catabolism in the tumor stroma and anabolic tumor cell expansion.
Figures



Comment in
-
Glutamine and the tumor microenvironment: understanding the mechanisms that fuel cancer progression.Cancer Biol Ther. 2011 Dec 15;12(12):1098-100. doi: 10.4161/cbt.12.12.18856. Epub 2011 Dec 15. Cancer Biol Ther. 2011. PMID: 22236873 Free PMC article. No abstract available.
-
Complexity of metabolic cancer control: can we exploit the superior metabolic position of glucose?Cancer Biol Ther. 2012 Jun;13(8):585. doi: 10.4161/cbt.20085. Epub 2012 Jun 1. Cancer Biol Ther. 2012. PMID: 22549159 No abstract available.
References
-
- Bode BP, Fuchs BC, Hurley BP, Conroy JL, Suetterlin JE, Tanabe KK, et al. Molecular and functional analysis of glutamine uptake in human hepatoma and liver-derived cells. Am J Physiol Gastrointest Liver Physiol. 2002;283:G1062–G1073. - PubMed
-
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA075503/CA/NCI NIH HHS/United States
- R01 CA098779/CA/NCI NIH HHS/United States
- R01-CA-120876/CA/NCI NIH HHS/United States
- R01 CA120876/CA/NCI NIH HHS/United States
- R01-CA-70896/CA/NCI NIH HHS/United States
- R01-CA-098779/CA/NCI NIH HHS/United States
- R01-CA-86072/CA/NCI NIH HHS/United States
- R01-AR-055660/AR/NIAMS NIH HHS/United States
- R01-CA-080250/CA/NCI NIH HHS/United States
- R01 CA070896/CA/NCI NIH HHS/United States
- P30 CA056036/CA/NCI NIH HHS/United States
- P30-CA-56036/CA/NCI NIH HHS/United States
- R01-CA-107382/CA/NCI NIH HHS/United States
- R01 AR055660/AR/NIAMS NIH HHS/United States
- R01-CA-75503/CA/NCI NIH HHS/United States
- R01 CA080250/CA/NCI NIH HHS/United States
- R01 CA086072/CA/NCI NIH HHS/United States
- K08 CA175193/CA/NCI NIH HHS/United States
- R01 CA107382/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
