Complete subunit architecture of the proteasome regulatory particle
- PMID: 22237024
- PMCID: PMC3285539
- DOI: 10.1038/nature10774
Complete subunit architecture of the proteasome regulatory particle
Abstract
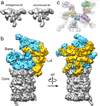
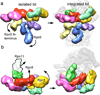
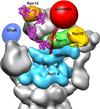
The proteasome is the major ATP-dependent protease in eukaryotic cells, but limited structural information restricts a mechanistic understanding of its activities. The proteasome regulatory particle, consisting of the lid and base subcomplexes, recognizes and processes polyubiquitinated substrates. Here we used electron microscopy and a new heterologous expression system for the lid to delineate the complete subunit architecture of the regulatory particle from yeast. Our studies reveal the spatial arrangement of ubiquitin receptors, deubiquitinating enzymes and the protein unfolding machinery at subnanometre resolution, outlining the substrate's path to degradation. Unexpectedly, the ATPase subunits within the base unfoldase are arranged in a spiral staircase, providing insight into potential mechanisms for substrate translocation through the central pore. Large conformational rearrangements of the lid upon holoenzyme formation suggest allosteric regulation of deubiquitination. We provide a structural basis for the ability of the proteasome to degrade a diverse set of substrates and thus regulate vital cellular processes.
Figures


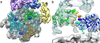
Comment in
-
Cell biology: Destruction deconstructed.Nature. 2012 Feb 8;482(7384):170-1. doi: 10.1038/482170a. Nature. 2012. PMID: 22318600 No abstract available.
References
-
- Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. - PubMed
References for full methods
-
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. - PubMed
-
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. - PubMed
-
- Sorzano CO, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

