Synthetic in vivo validation of gene network circuitry
- PMID: 22238426
- PMCID: PMC3277137
- DOI: 10.1073/pnas.1119905109
Synthetic in vivo validation of gene network circuitry
Abstract
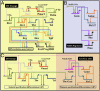
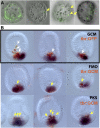
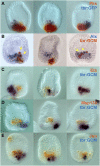
Embryonic development is controlled by networks of interacting regulatory genes. The individual linkages of gene regulatory networks (GRNs) are customarily validated by functional cis-regulatory analysis, but an additional approach to validation is to rewire GRN circuitry to test experimentally predictions derived from network structure. Here we use this synthetic method to challenge specific predictions of the sea urchin embryo endomesoderm GRN. Expression vectors generated by in vitro recombination of exogenous sequences into BACs were used to cause elements of a nonskeletogenic mesoderm GRN to be deployed in skeletogenic cells and to detect their effects. The result of reengineering the regulatory circuitry in this way was to divert the developmental program of these cells from skeletogenesis to pigment cell formation, confirming a direct prediction of the GRN. In addition, the experiment revealed previously undetected cryptic repression functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Sherwood DR, McClay DR. LvNotch signaling mediates secondary mesenchyme specification in the sea urchin embryo. Development. 1999;126:1703–1713. - PubMed
-
- Sweet HC, Hodor PG, Ettensohn CA. The role of micromere signaling in Notch activation and mesoderm specification during sea urchin embryogenesis. Development. 1999;126:5255–5265. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

