Evidence for transcript networks composed of chimeric RNAs in human cells
- PMID: 22238572
- PMCID: PMC3251577
- DOI: 10.1371/journal.pone.0028213
Evidence for transcript networks composed of chimeric RNAs in human cells
Abstract
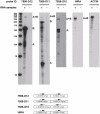
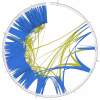
The classic organization of a gene structure has followed the Jacob and Monod bacterial gene model proposed more than 50 years ago. Since then, empirical determinations of the complexity of the transcriptomes found in yeast to human has blurred the definition and physical boundaries of genes. Using multiple analysis approaches we have characterized individual gene boundaries mapping on human chromosomes 21 and 22. Analyses of the locations of the 5' and 3' transcriptional termini of 492 protein coding genes revealed that for 85% of these genes the boundaries extend beyond the current annotated termini, most often connecting with exons of transcripts from other well annotated genes. The biological and evolutionary importance of these chimeric transcripts is underscored by (1) the non-random interconnections of genes involved, (2) the greater phylogenetic depth of the genes involved in many chimeric interactions, (3) the coordination of the expression of connected genes and (4) the close in vivo and three dimensional proximity of the genomic regions being transcribed and contributing to parts of the chimeric RNAs. The non-random nature of the connection of the genes involved suggest that chimeric transcripts should not be studied in isolation, but together, as an RNA network.
Conflict of interest statement
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

