Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus
- PMID: 22238576
- PMCID: PMC3253072
- DOI: 10.1371/journal.pone.0028316
Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus
Abstract
Background: Daptomycin remains one of our last-line anti-staphylococcal agents. This study aims to characterize the genetic evolution to daptomycin resistance in S. aureus.
Methods: Whole genome sequencing was performed on a unique collection of isogenic, clinical (21 strains) and laboratory (12 strains) derived strains that had been exposed to daptomycin and developed daptomycin-nonsusceptibility. Electron microscopy (EM) and lipid membrane studies were performed on selected isolates.
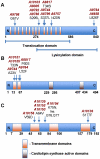
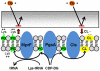
Results: On average, six coding region mutations were observed across the genome in the clinical daptomycin exposed strains, whereas only two mutations on average were seen in the laboratory exposed pairs. All daptomycin-nonsusceptible strains had a mutation in a phospholipid biosynthesis gene. This included mutations in the previously described mprF gene, but also in other phospholipid biosynthesis genes, including cardiolipin synthase (cls2) and CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase (pgsA). EM and lipid membrane composition analyses on two clinical pairs showed that the daptomycin-nonsusceptible strains had a thicker cell wall and an increase in membrane lysyl-phosphatidylglycerol.
Conclusion: Point mutations in genes coding for membrane phospholipids are associated with the development of reduced susceptibility to daptomycin in S. aureus. Mutations in cls2 and pgsA appear to be new genetic mechanisms affecting daptomycin susceptibility in S. aureus.
Conflict of interest statement
Figures


References
-
- Fowler VG, Jr, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med. 2006;355:653–665. - PubMed
-
- Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 2010;23:99–139. - PMC - PubMed
-
- Jung D, Powers JP, Straus SK, Hancock RE. Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem Phys Lipids. 2008;154:120–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

