Mutational analyses of the influenza A virus polymerase subunit PA reveal distinct functions related and unrelated to RNA polymerase activity
- PMID: 22238617
- PMCID: PMC3253111
- DOI: 10.1371/journal.pone.0029485
Mutational analyses of the influenza A virus polymerase subunit PA reveal distinct functions related and unrelated to RNA polymerase activity
Abstract
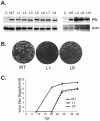
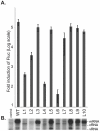
Influenza A viral polymerase is a heterotrimeric complex that consists of PA, PB1, and PB2 subunits. We previously reported that a di-codon substitution mutation (G507A-R508A), denoted J10, in the C-terminal half of PA had no apparent effect on viral RNA synthesis but prevented infectious virus production, indicating that PA may have a novel role independent of its polymerase activity. To further examine the roles of PA in the viral life cycle, we have now generated and characterized additional mutations in regions flanking the J10 site from residues 497 to 518. All tested di-codon mutations completely abolished or significantly reduced viral infectivity, but they did so through disparate mechanisms. Several showed effects resembling those of J10, in that the mutant polymerase supported normal levels of viral RNA synthesis but nonetheless failed to generate infectious viral particles. Others eliminated polymerase activity, in most cases by perturbing the normal nuclear localization of PA protein in cells. We also engineered single-codon mutations that were predicted to pack near the J10 site in the crystal structure of PA, and found that altering residues K378 or D478 each produced a J10-like phenotype. In further studies of J10 itself, we found that this mutation does not affect the formation and release of virion-like particles per se, but instead impairs the ability of those particles to incorporate each of the eight essential RNA segments (vRNAs) that make up the viral genome. Taken together, our analysis identifies mutations in the C-terminal region of PA that differentially affect at least three distinct activities: protein nuclear localization, viral RNA synthesis, and a trans-acting function that is required for efficient packaging of all eight vRNAs.
Conflict of interest statement
Figures

Similar articles
-
Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA.J Virol. 2006 Jan;80(1):252-61. doi: 10.1128/JVI.80.1.252-261.2006. J Virol. 2006. PMID: 16352550 Free PMC article.
-
Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments.J Virol. 2008 Jan;82(1):229-36. doi: 10.1128/JVI.01541-07. Epub 2007 Oct 24. J Virol. 2008. PMID: 17959657 Free PMC article.
-
Differential role of the influenza A virus polymerase PA subunit for vRNA and cRNA promoter binding.Virology. 2008 Jan 5;370(1):194-204. doi: 10.1016/j.virol.2007.08.029. Epub 2007 Oct 1. Virology. 2008. PMID: 17905403
-
The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication.Acta Virol. 2013;57(2):113-22. doi: 10.4149/av_2013_02_113. Acta Virol. 2013. PMID: 23600869 Review.
-
PA and PA-X: two key proteins from segment 3 of the influenza viruses.Front Cell Infect Microbiol. 2025 Mar 14;15:1560250. doi: 10.3389/fcimb.2025.1560250. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40160474 Free PMC article. Review.
Cited by
-
Duck innate immune responses to high and low pathogenicity H5 avian influenza viruses.Vet Microbiol. 2019 Jan;228:101-111. doi: 10.1016/j.vetmic.2018.11.018. Epub 2018 Nov 19. Vet Microbiol. 2019. PMID: 30593354 Free PMC article.
-
Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality.PLoS Genet. 2015 Jul 1;11(7):e1005310. doi: 10.1371/journal.pgen.1005310. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26132554 Free PMC article.
-
Residues in the PB2 and PA genes contribute to the pathogenicity of avian H7N3 influenza A virus in DBA/2 mice.Virology. 2016 Jul;494:89-99. doi: 10.1016/j.virol.2016.04.013. Epub 2016 Apr 20. Virology. 2016. PMID: 27105450 Free PMC article.
-
Molecular Evolution of Influenza A Viruses From Mauritius, 2017-2019.Influenza Other Respir Viruses. 2025 May;19(5):e70108. doi: 10.1111/irv.70108. Influenza Other Respir Viruses. 2025. PMID: 40360235 Free PMC article.
References
-
- Lamb RA, Krug RM. Orthomyxoviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields virology. 4th ed. Philadelphia, PA: Lippincott-Raven Publishers; 2003. pp. 1487–1531.
-
- Murti KG, Webster RG, Jones IM. Localization of RNA polymerases on influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

