Interaction of I50V mutant and I50L/A71V double mutant HIV-protease with inhibitor TMC114 (darunavir): molecular dynamics simulation and binding free energy studies
- PMID: 22239286
- PMCID: PMC3288396
- DOI: 10.1021/jp2074804
Interaction of I50V mutant and I50L/A71V double mutant HIV-protease with inhibitor TMC114 (darunavir): molecular dynamics simulation and binding free energy studies
Abstract
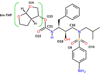
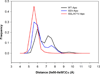
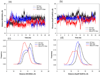
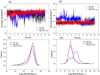
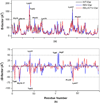
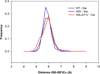
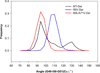
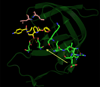
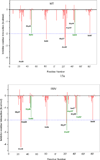
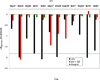
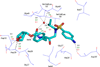
In the present work, the binding of inhibitor TMC114 (darunavir) to wild-type (WT), single (I50V) as well as double (I50L/A71V) mutant HIV-proteases (HIV-pr) was investigated with all-atom molecular dynamics (MD) simulations as well as molecular mechanic-Poisson-Boltzmann surface area (MM-PBSA) calculation. For both the apo and complexed HIV-pr, many intriguing effects due to double mutant, I50L/A71V, are observed. For example, the flap-flap distance and the distance from the active site to the flap residues in the apo I50L/A71V-HIV-pr are smaller than those of WT- and I50V-HIV-pr, probably making the active site smaller in volume and closer movement of flaps. For the complexed HIV-pr with TMC114, the double mutant I50L/A71V shows a less curling of the flap tips and less flexibility than WT and the single mutant I50V. As for the other previous studies, the present results also show that the single mutant I50V decreases the binding affinity of I50V-HIV-pr to TMC, resulting in a drug resistance; whereas the double mutant I50L/A71V increases the binding affinity, and as a result of the stronger binding, the I50L/A71V may be well adapted by the TMC114. The energy decomposition analysis suggests that the increase of the binding for the double mutant I50L/A71V-HIV-pr can be mainly attributed to the increase in electrostatic energy by -5.52 kacl/mol and van der Waals by -0.42 kcal/mol, which are canceled out in part by the increase of polar solvation energy of 1.99 kcal/mol. The I50L/A71V mutant directly increases the binding affinity by approximately -0.88 (Ile50 to Leu50) and -0.90 (Ile50' to Leu50') kcal/mol, accounting 45% for the total gain of the binding affinity. Besides the direct effects from the residues Leu50 and Leu50', the residue Gly49' increases the binding affinity of I50L/A71V-HIV-pr to the inhibitor by -0.74 kcal/mol, to which the electrostatic interaction of Leu50's backbone contributes by -1.23 kcal/mol. Another two residues Ile84 and Ile47' also increase the binding affinity by -0.22 and -0.29 kcal/mol, respectively, which can be mainly attributed to van der Waals terms (ΔT(vdw) = -0.21 and -0.39 kcal/mol).
Figures
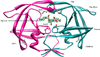







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

