Mutations in LPL, APOC2, APOA5, GPIHBP1 and LMF1 in patients with severe hypertriglyceridaemia
- PMID: 22239554
- PMCID: PMC3940136
- DOI: 10.1111/j.1365-2796.2012.02516.x
Mutations in LPL, APOC2, APOA5, GPIHBP1 and LMF1 in patients with severe hypertriglyceridaemia
Abstract
Objectives: The severe forms of hypertriglyceridaemia (HTG) are caused by mutations in genes that lead to the loss of function of lipoprotein lipase (LPL). In most patients with severe HTG (TG > 10 mmol L(-1) ), it is a challenge to define the underlying cause. We investigated the molecular basis of severe HTG in patients referred to the Lipid Clinic at the Academic Medical Center Amsterdam.
Methods: The coding regions of LPL, APOC2, APOA5 and two novel genes, lipase maturation factor 1 (LMF1) and GPI-anchored high-density lipoprotein (HDL)-binding protein 1 (GPIHBP1), were sequenced in 86 patients with type 1 and type 5 HTG and 327 controls.
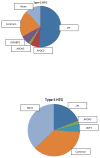
Results: In 46 patients (54%), rare DNA sequence variants were identified, comprising variants in LPL (n = 19), APOC2 (n = 1), APOA5 (n = 2), GPIHBP1 (n = 3) and LMF1 (n = 8). In 22 patients (26%), only common variants in LPL (p.Asp36Asn, p.Asn318Ser and p.Ser474Ter) and APOA5 (p.Ser19Trp) could be identified, whereas no mutations were found in 18 patients (21%). In vitro validation revealed that the mutations in LMF1 were not associated with compromised LPL function. Consistent with this, five of the eight LMF1 variants were also found in controls and therefore cannot account for the observed phenotype.
Conclusions: The prevalence of mutations in LPL was 34% and mostly restricted to patients with type 1 HTG. Mutations in GPIHBP1 (n = 3), APOC2 (n = 1) and APOA5 (n = 2) were rare but the associated clinical phenotype was severe. Routine sequencing of candidate genes in severe HTG has improved our understanding of the molecular basis of this phenotype associated with acute pancreatitis and may help to guide future individualized therapeutic strategies.
© 2012 The Association for the Publication of the Journal of Internal Medicine.
Conflict of interest statement
Figures

References
-
- de Graaf J, Couture P, Sniderman A. A diagnostic algorithm for the atherogenic apolipoprotein B dyslipoproteinemias. Nat Clin Pract Endocrinol Metab. 2008;4:608–18. - PubMed
-
- Avis HJ, Scheffer HJ, Kastelein JJ, Dallinga-Thie GM, Wijburg FA. Pink-creamy whole blood in a 3-month-old infant with a homozygous deletion in the lipoprotein lipase gene. Clin Genet. 2010;77:430–3. - PubMed
-
- Rahalkar AR, Hegele RA. Monogenic pediatric dyslipidemias: classification, genetics and clinical spectrum. Mol Genet Metab. 2008;93:282–94. - PubMed
-
- Brunzell JD. Familial Lipoprotein Lipase Deficiency and other causes of Chylomicronemia Syndrome. In: Scriver ABA, Sly W, Valle D, editors. The Molecular Basis of Inherited Disease. New York: McGraw Hill; 1995. pp. 1913–32.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

