Lim homeobox genes in the Ctenophore Mnemiopsis leidyi: the evolution of neural cell type specification
- PMID: 22239757
- PMCID: PMC3283466
- DOI: 10.1186/2041-9139-3-2
Lim homeobox genes in the Ctenophore Mnemiopsis leidyi: the evolution of neural cell type specification
Abstract
Background: Nervous systems are thought to be important to the evolutionary success and diversification of metazoans, yet little is known about the origin of simple nervous systems at the base of the animal tree. Recent data suggest that ctenophores, a group of macroscopic pelagic marine invertebrates, are the most ancient group of animals that possess a definitive nervous system consisting of a distributed nerve net and an apical statocyst. This study reports on details of the evolution of the neural cell type specifying transcription factor family of LIM homeobox containing genes (Lhx), which have highly conserved functions in neural specification in bilaterian animals.
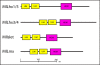
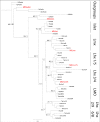
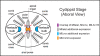
Results: Using next generation sequencing, the first draft of the genome of the ctenophore Mnemiopsis leidyi has been generated. The Lhx genes in all animals are represented by seven subfamilies (Lhx1/5, Lhx3/4, Lmx, Islet, Lhx2/9, Lhx6/8, and LMO) of which four were found to be represented in the ctenophore lineage (Lhx1/5, Lhx3/4, Lmx, and Islet). Interestingly, the ctenophore Lhx gene complement is more similar to the sponge complement (sponges do not possess neurons) than to either the cnidarian-bilaterian or placozoan Lhx complements. Using whole mount in situ hybridization, the Lhx gene expression patterns were examined and found to be expressed around the blastopore and in cells that give rise to the apical organ and putative neural sensory cells.
Conclusion: This research gives us a first look at neural cell type specification in the ctenophore M. leidyi. Within M. leidyi, Lhx genes are expressed in overlapping domains within proposed neural cellular and sensory cell territories. These data suggest that Lhx genes likely played a conserved role in the patterning of sensory cells in the ancestor of sponges and ctenophores, and may provide a link to the expression of Lhx orthologs in sponge larval photoreceptive cells. Lhx genes were later co-opted into patterning more diversified complements of neural and non-neural cell types in later evolving animals.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

