Direct interaction of ligand-receptor pairs specifying stomatal patterning
- PMID: 22241782
- PMCID: PMC3273837
- DOI: 10.1101/gad.179895.111
Direct interaction of ligand-receptor pairs specifying stomatal patterning
Abstract
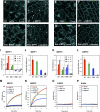
Valves on the plant epidermis called stomata develop according to positional cues, which likely involve putative ligands (EPIDERMAL PATTERNING FACTORS [EPFs]) and putative receptors (ERECTA family receptor kinases and TOO MANY MOUTHS [TMM]) in Arabidopsis. Here we report the direct, robust, and saturable binding of bioactive EPF peptides to the ERECTA family. In contrast, TMM exhibits negligible binding to EPF1 but binding to EPF2. The ERECTA family forms receptor homomers in vivo. On the other hand, TMM associates with the ERECTA family but not with itself. While ERECTA family receptor kinases exhibit complex redundancy, blocking ERECTA and ERECTA-LIKE1 (ERL1) signaling confers specific insensitivity to EPF2 and EPF1, respectively. Our results place the ERECTA family as the primary receptors for EPFs with TMM as a signal modulator and establish EPF2-ERECTA and EPF1-ERL1 as ligand-receptor pairs specifying two steps of stomatal development: initiation and spacing divisions.
Figures


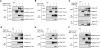

References
-
- Abrash EB, Bergmann DC 2010. Regional specification of stomatal production by the putative ligand CHALLAH. Development 137: 447–455 - PubMed
-
- Baltus RE, Carmon KS, Luck LA 2007. Quartz crystal microbalance (QCM) with immobilized protein receptors: Comparison of response to ligand binding for direct protein immobilization and protein attachment via disulfide linker. Langmuir 23: 3880–3885 - PubMed
-
- Belkhadir Y, Chory J 2006. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 - PubMed
-
- Clark SE, Williams RW, Meyerowitz EM 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
