Identification and characterization of a novel non-structural protein of bluetongue virus
- PMID: 22241985
- PMCID: PMC3248566
- DOI: 10.1371/journal.ppat.1002477
Identification and characterization of a novel non-structural protein of bluetongue virus
Abstract
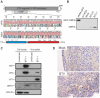
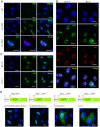
Bluetongue virus (BTV) is the causative agent of a major disease of livestock (bluetongue). For over two decades, it has been widely accepted that the 10 segments of the dsRNA genome of BTV encode for 7 structural and 3 non-structural proteins. The non-structural proteins (NS1, NS2, NS3/NS3a) play different key roles during the viral replication cycle. In this study we show that BTV expresses a fourth non-structural protein (that we designated NS4) encoded by an open reading frame in segment 9 overlapping the open reading frame encoding VP6. NS4 is 77-79 amino acid residues in length and highly conserved among several BTV serotypes/strains. NS4 was expressed early post-infection and localized in the nucleoli of BTV infected cells. By reverse genetics, we showed that NS4 is dispensable for BTV replication in vitro, both in mammalian and insect cells, and does not affect viral virulence in murine models of bluetongue infection. Interestingly, NS4 conferred a replication advantage to BTV-8, but not to BTV-1, in cells in an interferon (IFN)-induced antiviral state. However, the BTV-1 NS4 conferred a replication advantage both to a BTV-8 reassortant containing the entire segment 9 of BTV-1 and to a BTV-8 mutant with the NS4 identical to the homologous BTV-1 protein. Collectively, this study suggests that NS4 plays an important role in virus-host interaction and is one of the mechanisms played, at least by BTV-8, to counteract the antiviral response of the host. In addition, the distinct nucleolar localization of NS4, being expressed by a virus that replicates exclusively in the cytoplasm, offers new avenues to investigate the multiple roles played by the nucleolus in the biology of the cell.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Mellor PS, Baylis M, Mertens PP. London: Academic Press; 2009. Bluetongue; Pastoret P-P, editor.
-
- Maclachlan NJ, Drew CP, Darpel KE, Worwa G. The pathology and pathogenesis of bluetongue. J Comp Pathol. 2009;141:1–16. - PubMed
-
- Schwartz-Cornil I, Mertens PP, Contreras V, Hemati B, Pascale F, et al. Bluetongue virus: virology, pathogenesis and immunity. Vet Res. 2008;39:46. - PubMed
-
- Erasmus BJ, Potgieter AC. The history of blueotngue. In: Mellor PS, Baylis M, Mertens P, editors. Bluetongue. Amsterdam: Elsevier; 2009. pp. 7–21.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

