Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma
- PMID: 22241995
- PMCID: PMC3252369
- DOI: 10.1371/journal.ppat.1002470
Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma
Abstract
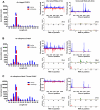
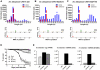
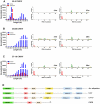
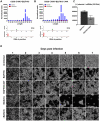
The natural maintenance cycles of many mosquito-borne pathogens require establishment of persistent non-lethal infections in the invertebrate host. The mechanism by which this occurs is not well understood, but we have previously shown that an antiviral response directed by small interfering RNAs (siRNAs) is important in modulating the pathogenesis of alphavirus infections in the mosquito. However, we report here that infection of mosquitoes with an alphavirus also triggers the production of another class of virus-derived small RNAs that exhibit many similarities to ping-pong-dependent piwi-interacting RNAs (piRNAs). However, unlike ping-pong-dependent piRNAs that have been described previously from repetitive elements or piRNA clusters, our work suggests production in the soma. We also present evidence that suggests virus-derived piRNA-like small RNAs are capable of modulating the pathogenesis of alphavirus infections in dicer-2 null mutant mosquito cell lines defective in viral siRNA production. Overall, our results suggest that a non-canonical piRNA pathway is present in the soma of vector mosquitoes and may be acting redundantly to the siRNA pathway to target alphavirus replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

