The Caenorhabditis elegans synthetic multivulva genes prevent ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF
- PMID: 22242000
- PMCID: PMC3248470
- DOI: 10.1371/journal.pgen.1002418
The Caenorhabditis elegans synthetic multivulva genes prevent ras pathway activation by tightly repressing global ectopic expression of lin-3 EGF
Abstract
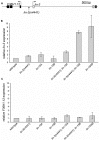
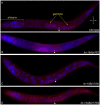
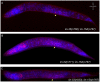
The Caenorhabditis elegans class A and B synthetic multivulva (synMuv) genes redundantly antagonize an EGF/Ras pathway to prevent ectopic vulval induction. We identify a class A synMuv mutation in the promoter of the lin-3 EGF gene, establishing that lin-3 is the key biological target of the class A synMuv genes in vulval development and that the repressive activities of the class A and B synMuv pathways are integrated at the level of lin-3 expression. Using FISH with single mRNA molecule resolution, we find that lin-3 EGF expression is tightly restricted to only a few tissues in wild-type animals, including the germline. In synMuv double mutants, lin-3 EGF is ectopically expressed at low levels throughout the animal. Our findings reveal that the widespread ectopic expression of a growth factor mRNA at concentrations much lower than that in the normal domain of expression can abnormally activate the Ras pathway and alter cell fates. These results suggest hypotheses for the mechanistic basis of the functional redundancy between the tumor-suppressor-like class A and B synMuv genes: the class A synMuv genes either directly or indirectly specifically repress ectopic lin-3 expression; while the class B synMuv genes might function similarly, but alternatively might act to repress lin-3 as a consequence of their role in preventing cells from adopting a germline-like fate. Analogous genes in mammals might function as tumor suppressors by preventing broad ectopic expression of EGF-like ligands.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The LIN-15A and LIN-56 transcriptional regulators interact to negatively regulate EGF/Ras signaling in Caenorhabditis elegans vulval cell-fate determination.Genetics. 2011 Mar;187(3):803-15. doi: 10.1534/genetics.110.124487. Epub 2010 Dec 31. Genetics. 2011. PMID: 21196525 Free PMC article.
-
SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans.Dev Cell. 2006 May;10(5):667-72. doi: 10.1016/j.devcel.2006.04.001. Dev Cell. 2006. PMID: 16678779
-
Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development.Genetics. 2006 Jun;173(2):709-26. doi: 10.1534/genetics.106.056465. Epub 2006 Apr 19. Genetics. 2006. PMID: 16624904 Free PMC article.
-
LET-23-mediated signal transduction during Caenorhabditis elegans development.Mol Reprod Dev. 1995 Dec;42(4):523-8. doi: 10.1002/mrd.1080420422. Mol Reprod Dev. 1995. PMID: 8607985 Review.
-
Vulval development.WormBook. 2005 Jun 25:1-28. doi: 10.1895/wormbook.1.6.1. WormBook. 2005. PMID: 18050418 Free PMC article. Review.
Cited by
-
Decreased SynMuv B gene activity in response to viral infection leads to activation of the antiviral RNAi pathway in C. elegans.PLoS Biol. 2025 Jan 29;23(1):e3002748. doi: 10.1371/journal.pbio.3002748. eCollection 2025 Jan. PLoS Biol. 2025. PMID: 39879188 Free PMC article.
-
Caenorhabditis elegans SynMuv B gene activity is down-regulated during a viral infection to enhance RNA interference.bioRxiv [Preprint]. 2024 Jul 16:2024.07.12.603258. doi: 10.1101/2024.07.12.603258. bioRxiv. 2024. PMID: 39071373 Free PMC article. Preprint.
-
Reciprocal EGFR signaling in the anchor cell ensures precise inter-organ connection during Caenorhabditis elegans vulval morphogenesis.Development. 2022 Jan 1;149(1):dev199900. doi: 10.1242/dev.199900. Epub 2022 Jan 4. Development. 2022. PMID: 34982813 Free PMC article.
-
The Lipocalin LPR-1 Cooperates with LIN-3/EGF Signaling To Maintain Narrow Tube Integrity in Caenorhabditis elegans.Genetics. 2017 Mar;205(3):1247-1260. doi: 10.1534/genetics.116.195156. Epub 2016 Dec 30. Genetics. 2017. PMID: 28040739 Free PMC article.
-
Single-molecule methods for studying gene regulation in vivo.Pflugers Arch. 2013 Mar;465(3):383-95. doi: 10.1007/s00424-013-1243-y. Epub 2013 Feb 21. Pflugers Arch. 2013. PMID: 23430319 Free PMC article. Review.
References
-
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. - PubMed
-
- Massague J. Transforming growth factor-alpha. A model for membrane-anchored growth factors. J Biol Chem. 1990;265:21393–21396. - PubMed
-
- Yotsumoto F, Yagi H, Suzuki SO, Oki E, Tsujioka H, et al. Validation of HB-EGF and amphiregulin as targets for human cancer therapy. Biochem Biophys Res Commun. 2008;365:555–561. - PubMed
-
- Jhappan C, Stahle C, Harkins RN, Fausto N, Smith GH, et al. TGF alpha overexpression in transgenic mice induces liver neoplasia and abnormal development of the mammary gland and pancreas. Cell. 1990;61:1137–1146. - PubMed
-
- Sandgren EP, Luetteke NC, Palmiter RD, Brinster RL, Lee DC. Overexpression of TGF alpha in transgenic mice: induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell. 1990;61:1121–1135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

