MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila
- PMID: 22242005
- PMCID: PMC3248469
- DOI: 10.1371/journal.pgen.1002429
MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila
Abstract
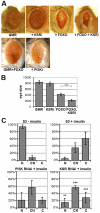
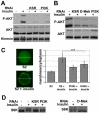
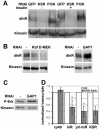
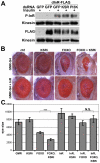
The insulin/IGF-activated AKT signaling pathway plays a crucial role in regulating tissue growth and metabolism in multicellular animals. Although core components of the pathway are well defined, less is known about mechanisms that adjust the sensitivity of the pathway to extracellular stimuli. In humans, disturbance in insulin sensitivity leads to impaired clearance of glucose from the blood stream, which is a hallmark of diabetes. Here we present the results of a genetic screen in Drosophila designed to identify regulators of insulin sensitivity in vivo. Components of the MAPK/ERK pathway were identified as modifiers of cellular insulin responsiveness. Insulin resistance was due to downregulation of insulin-like receptor gene expression following persistent MAPK/ERK inhibition. The MAPK/ERK pathway acts via the ETS-1 transcription factor Pointed. This mechanism permits physiological adjustment of insulin sensitivity and subsequent maintenance of circulating glucose at appropriate levels.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila.Nature. 2001 May 17;411(6835):330-4. doi: 10.1038/35077122. Nature. 2001. PMID: 11357138
-
Lessons from Drosophila Pointed, an ETS family transcription factor and key nuclear effector of the RTK signaling pathway.Genesis. 2018 Dec;56(11-12):e23257. doi: 10.1002/dvg.23257. Epub 2018 Nov 25. Genesis. 2018. PMID: 30318758 Review.
-
MAE, a dual regulator of the EGFR signaling pathway, is a target of the Ets transcription factors PNT and YAN.Mech Dev. 2004 Dec;121(12):1469-79. doi: 10.1016/j.mod.2004.07.009. Mech Dev. 2004. PMID: 15511639
-
Neuronal Cbl controls biosynthesis of insulin-like peptides in Drosophila melanogaster.Mol Cell Biol. 2012 Sep;32(18):3610-23. doi: 10.1128/MCB.00592-12. Epub 2012 Jul 9. Mol Cell Biol. 2012. PMID: 22778134 Free PMC article.
-
Understanding Forkhead box class O function: lessons from Drosophila melanogaster.Antioxid Redox Signal. 2011 Feb 15;14(4):635-47. doi: 10.1089/ars.2010.3407. Epub 2010 Oct 20. Antioxid Redox Signal. 2011. PMID: 20618068 Review.
Cited by
-
The different facets of heme-oxygenase 1 in innate and adaptive immunity.Cell Biochem Biophys. 2022 Dec;80(4):609-631. doi: 10.1007/s12013-022-01087-z. Epub 2022 Aug 26. Cell Biochem Biophys. 2022. PMID: 36018440 Review.
-
ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation.Genes Dev. 2013 Feb 15;27(4):441-9. doi: 10.1101/gad.201731.112. Genes Dev. 2013. PMID: 23431056 Free PMC article.
-
Methylglyoxal Induced Modifications to Stabilize Therapeutic Proteins: A Review.Protein J. 2024 Feb;43(1):39-47. doi: 10.1007/s10930-023-10166-w. Epub 2023 Nov 28. Protein J. 2024. PMID: 38017314 Review.
-
NLRP3 inflammasome via IL-1β regulates PCSK9 secretion.Theranostics. 2020 May 30;10(16):7100-7110. doi: 10.7150/thno.45939. eCollection 2020. Theranostics. 2020. PMID: 32641981 Free PMC article.
-
Genetic variation of macronutrient tolerance in Drosophila melanogaster.Nat Commun. 2022 Mar 28;13(1):1637. doi: 10.1038/s41467-022-29183-x. Nat Commun. 2022. PMID: 35347148 Free PMC article.
References
-
- Hietakangas V, Cohen SM. Regulation of tissue growth through nutrient sensing. Annu Rev Genet. 2009;43:389–410. - PubMed
-
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. - PubMed
-
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. - PubMed
-
- Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–40416. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

