The RNA silencing enzyme RNA polymerase v is required for plant immunity
- PMID: 22242006
- PMCID: PMC3248562
- DOI: 10.1371/journal.pgen.1002434
The RNA silencing enzyme RNA polymerase v is required for plant immunity
Abstract
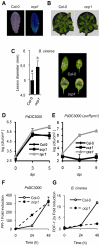
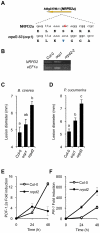
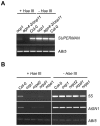
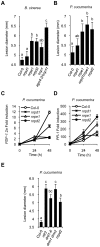
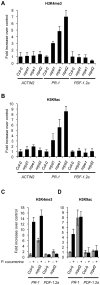
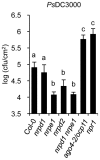
RNA-directed DNA methylation (RdDM) is an epigenetic control mechanism driven by small interfering RNAs (siRNAs) that influence gene function. In plants, little is known of the involvement of the RdDM pathway in regulating traits related to immune responses. In a genetic screen designed to reveal factors regulating immunity in Arabidopsis thaliana, we identified NRPD2 as the OVEREXPRESSOR OF CATIONIC PEROXIDASE 1 (OCP1). NRPD2 encodes the second largest subunit of the plant-specific RNA Polymerases IV and V (Pol IV and Pol V), which are crucial for the RdDM pathway. The ocp1 and nrpd2 mutants showed increases in disease susceptibility when confronted with the necrotrophic fungal pathogens Botrytis cinerea and Plectosphaerella cucumerina. Studies were extended to other mutants affected in different steps of the RdDM pathway, such as nrpd1, nrpe1, ago4, drd1, rdr2, and drm1drm2 mutants. Our results indicate that all the mutants studied, with the exception of nrpd1, phenocopy the nrpd2 mutants; and they suggest that, while Pol V complex is required for plant immunity, Pol IV appears dispensable. Moreover, Pol V defective mutants, but not Pol IV mutants, show enhanced disease resistance towards the bacterial pathogen Pseudomonas syringae DC3000. Interestingly, salicylic acid (SA)-mediated defenses effective against PsDC3000 are enhanced in Pol V defective mutants, whereas jasmonic acid (JA)-mediated defenses that protect against fungi are reduced. Chromatin immunoprecipitation analysis revealed that, through differential histone modifications, SA-related defense genes are poised for enhanced activation in Pol V defective mutants and provide clues for understanding the regulation of gene priming during defense. Our results highlight the importance of epigenetic control as an additional layer of complexity in the regulation of plant immunity and point towards multiple components of the RdDM pathway being involved in plant immunity based on genetic evidence, but whether this is a direct or indirect effect on disease-related genes is unclear.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009 [doi:10.1016/j.ceb.2009.01.025] - PubMed
-
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

