Common variants show predicted polygenic effects on height in the tails of the distribution, except in extremely short individuals
- PMID: 22242009
- PMCID: PMC3248463
- DOI: 10.1371/journal.pgen.1002439
Common variants show predicted polygenic effects on height in the tails of the distribution, except in extremely short individuals
Abstract
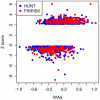
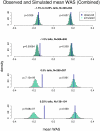
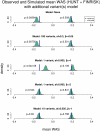
Common genetic variants have been shown to explain a fraction of the inherited variation for many common diseases and quantitative traits, including height, a classic polygenic trait. The extent to which common variation determines the phenotype of highly heritable traits such as height is uncertain, as is the extent to which common variation is relevant to individuals with more extreme phenotypes. To address these questions, we studied 1,214 individuals from the top and bottom extremes of the height distribution (tallest and shortest ∼1.5%), drawn from ∼78,000 individuals from the HUNT and FINRISK cohorts. We found that common variants still influence height at the extremes of the distribution: common variants (49/141) were nominally associated with height in the expected direction more often than is expected by chance (p<5×10⁻²⁸), and the odds ratios in the extreme samples were consistent with the effects estimated previously in population-based data. To examine more closely whether the common variants have the expected effects, we calculated a weighted allele score (WAS), which is a weighted prediction of height for each individual based on the previously estimated effect sizes of the common variants in the overall population. The average WAS is consistent with expectation in the tall individuals, but was not as extreme as expected in the shortest individuals (p<0.006), indicating that some of the short stature is explained by factors other than common genetic variation. The discrepancy was more pronounced (p<10⁻⁶) in the most extreme individuals (height<0.25 percentile). The results at the extreme short tails are consistent with a large number of models incorporating either rare genetic non-additive or rare non-genetic factors that decrease height. We conclude that common genetic variants are associated with height at the extremes as well as across the population, but that additional factors become more prominent at the shorter extreme.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Hundreds of variants clustered in genomic loci and biological pathways affect human height.Nature. 2010 Oct 14;467(7317):832-8. doi: 10.1038/nature09410. Epub 2010 Sep 29. Nature. 2010. PMID: 20881960 Free PMC article.
-
Rare and low-frequency coding variants alter human adult height.Nature. 2017 Feb 9;542(7640):186-190. doi: 10.1038/nature21039. Epub 2017 Feb 1. Nature. 2017. PMID: 28146470 Free PMC article.
-
Inference of the genetic architecture underlying BMI and height with the use of 20,240 sibling pairs.Am J Hum Genet. 2013 Nov 7;93(5):865-75. doi: 10.1016/j.ajhg.2013.10.005. Epub 2013 Oct 31. Am J Hum Genet. 2013. PMID: 24183453 Free PMC article.
-
Height matters-from monogenic disorders to normal variation.Nat Rev Endocrinol. 2013 Mar;9(3):171-7. doi: 10.1038/nrendo.2012.251. Epub 2013 Jan 22. Nat Rev Endocrinol. 2013. PMID: 23337954 Review.
-
Genetic regulation of adult stature.Curr Opin Pediatr. 2009 Aug;21(4):515-22. doi: 10.1097/MOP.0b013e32832c6dce. Curr Opin Pediatr. 2009. PMID: 19455035 Review.
Cited by
-
Identification and analysis of individuals who deviate from their genetically-predicted phenotype.PLoS Genet. 2023 Sep 21;19(9):e1010934. doi: 10.1371/journal.pgen.1010934. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37733769 Free PMC article.
-
Sibling similarity can reveal key insights into genetic architecture.Elife. 2025 Jan 8;12:RP87522. doi: 10.7554/eLife.87522. Elife. 2025. PMID: 39773384 Free PMC article.
-
Common DNA variants predict tall stature in Europeans.Hum Genet. 2014 May;133(5):587-97. doi: 10.1007/s00439-013-1394-0. Epub 2013 Nov 20. Hum Genet. 2014. PMID: 24253421
-
Genetic evaluation of short stature.J Clin Endocrinol Metab. 2014 Sep;99(9):3080-92. doi: 10.1210/jc.2014-1506. Epub 2014 Jun 10. J Clin Endocrinol Metab. 2014. PMID: 24915122 Free PMC article. Review.
-
Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture.Nat Genet. 2013 May;45(5):501-12. doi: 10.1038/ng.2606. Epub 2013 Apr 7. Nat Genet. 2013. PMID: 23563607 Free PMC article.
References
-
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

