The promoter of the cereal VERNALIZATION1 gene is sufficient for transcriptional induction by prolonged cold
- PMID: 22242122
- PMCID: PMC3248443
- DOI: 10.1371/journal.pone.0029456
The promoter of the cereal VERNALIZATION1 gene is sufficient for transcriptional induction by prolonged cold
Abstract
The VERNALIZATION1 (VRN1) gene of temperate cereals is transcriptionally activated by prolonged cold during winter (vernalization) to promote flowering. To investigate the mechanisms controlling induction of VRN1 by prolonged cold, different regions of the VRN1 gene were fused to the GREEN FLUORESCENT PROTEIN (GFP) reporter and expression of the resulting gene constructs was assayed in transgenic barley (Hordeum vulgare). A 2 kb segment of the promoter of VRN1 was sufficient for GFP expression in the leaves and shoot apex of transgenic barley plants. Fluorescence increased at the shoot apex prior to inflorescence initiation and was subsequently maintained in the developing inflorescence. The promoter was also sufficient for low-temperature induction of GFP expression. A naturally occurring insertion in the proximal promoter, which is associated with elevated VRN1 expression and early flowering in some spring wheats, did not abolish induction of VRN1 transcription by prolonged cold, however. A translational fusion of the promoter and transcribed regions of VRN1 to GFP, VRN1::GFP, was localised to nuclei of cells at the shoot apex of transgenic barley plants. The distribution of VRN1::GFP at the shoot apex was similar to the expression pattern of the VRN1 promoter-GFP reporter gene. Fluorescence from the VRN1::GFP fusion protein increased in the developing leaves after prolonged cold treatment. These observations suggest that the promoter of VRN1 is targeted by mechanisms that trigger vernalization-induced flowering in economically important temperate cereal crops.
Conflict of interest statement
Figures

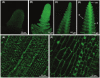




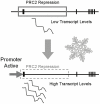
References
-
- Chouard P. Vernalization and its relation to dormancy. Ann Rev Plant Physiol. 1960;1:191–238.
-
- Trevaskis B. The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Functional Plant Biol. 2010;37:479–487.
-
- Trevaskis B, Hemming MN, Dennis ES, Peacock WJ. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007;12:352–357. - PubMed
-
- Distelfeld A, Li C, Dubcovsky J. Regulation of flowering in temperate cereals. Current Opinion Plant Biol. 2009;12:178–184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

