The tmRNA ribosome-rescue system
- PMID: 22243584
- PMCID: PMC3358797
- DOI: 10.1016/B978-0-12-386497-0.00005-0
The tmRNA ribosome-rescue system
Abstract
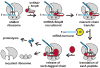
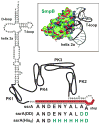
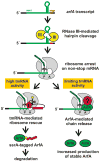
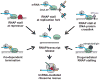
The bacterial tmRNA quality control system monitors protein synthesis and recycles stalled translation complexes in a process termed "ribosome rescue." During rescue, tmRNA acts first as a transfer RNA to bind stalled ribosomes, then as a messenger RNA to add the ssrA peptide tag to the C-terminus of the nascent polypeptide chain. The ssrA peptide targets tagged peptides for proteolysis, ensuring rapid degradation of potentially deleterious truncated polypeptides. Ribosome rescue also facilitates turnover of the damaged messages responsible for translational arrest. Thus, tmRNA increases the fidelity of gene expression by promoting the synthesis of full-length proteins. In addition to serving as a global quality control system, tmRNA also plays important roles in bacterial development, pathogenesis, and environmental stress responses. This review focuses on the mechanism of tmRNA-mediated ribosome rescue and the role of tmRNA in bacterial physiology.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
References
-
- Abe T, Sakaki K, Fujihara A, Ujiie H, Ushida C, Himeno H, Sato T, Muto A. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Molecular Microbiology. 2008;69:1491–1498. - PubMed
-
- Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes to Cells. 2002;7:629–638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

