Sequencing our way towards understanding global eukaryotic biodiversity
- PMID: 22244672
- PMCID: PMC3311718
- DOI: 10.1016/j.tree.2011.11.010
Sequencing our way towards understanding global eukaryotic biodiversity
Abstract
Microscopic eukaryotes are abundant, diverse and fill critical ecological roles across every ecosystem on Earth, yet there is a well-recognized gap in understanding of their global biodiversity. Fundamental advances in DNA sequencing and bioinformatics now allow accurate en masse biodiversity assessments of microscopic eukaryotes from environmental samples. Despite a promising outlook, the field of eukaryotic marker gene surveys faces significant challenges: how to generate data that are most useful to the community, especially in the face of evolving sequencing technologies and bioinformatics pipelines, and how to incorporate an expanding number of target genes.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

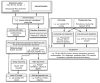
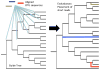
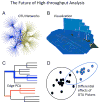

Comment in
-
Transcriptomics and microbial eukaryote diversity: a way forward.Trends Ecol Evol. 2012 Dec;27(12):651-2; author reply 652-3. doi: 10.1016/j.tree.2012.08.011. Epub 2012 Aug 31. Trends Ecol Evol. 2012. PMID: 22940223 No abstract available.
References
-
- Danovaro R, et al. Exponential decline of deep-sea ecosystem functioning linked to benthic biodiversity loss. Current Biology. 2008;18:1–18. - PubMed
-
- Wardle DA. The influence of biotic interactions on soil biodiversity. Ecology Letters. 2006;9:870–886. - PubMed
-
- Groisillier A, et al. Genetic diversity and habitats of two enigmatic marine alveolate lineages. Aquatic Microbial Ecology. 2006;42:277–291.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

