NMR structure of a heterodimeric SAM:SAM complex: characterization and manipulation of EphA2 binding reveal new cellular functions of SHIP2
- PMID: 22244754
- PMCID: PMC3516615
- DOI: 10.1016/j.str.2011.11.013
NMR structure of a heterodimeric SAM:SAM complex: characterization and manipulation of EphA2 binding reveal new cellular functions of SHIP2
Abstract
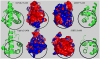
The sterile alpha motif (SAM) for protein-protein interactions is encountered in over 200 proteins, but the structural basis for its interactions is just becoming clear. Here we solved the structure of the EphA2-SHIP2 SAM:SAM heterodimeric complex by use of NMR restraints from chemical shift perturbations, NOE and RDC experiments. Specific contacts between the protein surfaces differ significantly from a previous model and other SAM:SAM complexes. Molecular dynamics and docking simulations indicate fluctuations in the complex toward alternate, higher energy conformations. The interface suggests that EphA family members bind to SHIP2 SAM, whereas EphB members may not; correspondingly, we demonstrate binding of EphA1, but not of EphB2, to SHIP2. A variant of EphB2 SAM was designed that binds SHIP2. Functional characterization of a mutant EphA2 compromised in SHIP2 binding reveals two previously unrecognized functions of SHIP2 in suppressing ligand-induced activation of EphA2 and in promoting receptor coordinated chemotactic cell migration.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures





References
-
- Baker JM, Candy DJ, Hawker RJ. Influences of pH on human platelet metabolism. Platelets. 2001;12:333–342. - PubMed
-
- Bhattacharjya S, Xu P, Gingras R, Shaykhutdinov R, Wu C, Whiteway M, Ni F. Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex. J Mol Biol. 2004;344:1071–1087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

