A DR4:tBID axis drives the p53 apoptotic response by promoting oligomerization of poised BAX
- PMID: 22246181
- PMCID: PMC3298004
- DOI: 10.1038/emboj.2011.498
A DR4:tBID axis drives the p53 apoptotic response by promoting oligomerization of poised BAX
Abstract
The cellular response to p53 activation varies greatly in a stimulus- and cell type-specific manner. Dissecting the molecular mechanisms defining these cell fate choices will assist the development of effective p53-based cancer therapies and also illuminate fundamental processes by which gene networks control cellular behaviour. Using an experimental system wherein stimulus-specific p53 responses are elicited by non-genotoxic versus genotoxic agents, we discovered a novel mechanism that determines whether cells undergo proliferation arrest or cell death. Strikingly, we observe that key mediators of cell-cycle arrest (p21, 14-3-3σ) and apoptosis (PUMA, BAX) are equally activated regardless of outcome. In fact, arresting cells display strong translocation of PUMA and BAX to the mitochondria, yet fail to release cytochrome C or activate caspases. Surprisingly, the key differential events in apoptotic cells are p53-dependent activation of the DR4 death receptor pathway, caspase 8-mediated cleavage of BID, and BID-dependent activation of poised BAX at the mitochondria. These results reveal a previously unappreciated role for DR4 and the extrinsic apoptotic pathway in cell fate choice following p53 activation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

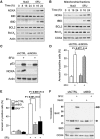
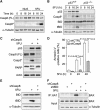
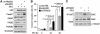



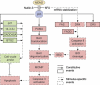
Similar articles
-
Bax/Bak activation in the absence of Bid, Bim, Puma, and p53.Cell Death Dis. 2016 Jun 16;7(6):e2266. doi: 10.1038/cddis.2016.167. Cell Death Dis. 2016. PMID: 27310874 Free PMC article.
-
Cleavage by Caspase 8 and Mitochondrial Membrane Association Activate the BH3-only Protein Bid during TRAIL-induced Apoptosis.J Biol Chem. 2016 May 27;291(22):11843-51. doi: 10.1074/jbc.M115.711051. Epub 2016 Apr 6. J Biol Chem. 2016. PMID: 27053107 Free PMC article.
-
Bcl-2 family member Bfl-1/A1 sequesters truncated bid to inhibit is collaboration with pro-apoptotic Bak or Bax.J Biol Chem. 2002 Jun 21;277(25):22781-8. doi: 10.1074/jbc.M201469200. Epub 2002 Apr 19. J Biol Chem. 2002. PMID: 11929871
-
Involvement of cardiolipin in tBID-induced activation of BAX during apoptosis.Chem Phys Lipids. 2014 Apr;179:70-4. doi: 10.1016/j.chemphyslip.2013.12.002. Epub 2013 Dec 12. Chem Phys Lipids. 2014. PMID: 24333953 Review.
-
Apoptosis - the p53 network.J Cell Sci. 2003 Oct 15;116(Pt 20):4077-85. doi: 10.1242/jcs.00739. J Cell Sci. 2003. PMID: 12972501 Review.
Cited by
-
The impact of post-transcriptional regulation in the p53 network.Brief Funct Genomics. 2013 Jan;12(1):46-57. doi: 10.1093/bfgp/els058. Epub 2012 Dec 14. Brief Funct Genomics. 2013. PMID: 23242178 Free PMC article. Review.
-
The NKL-code for innate lymphoid cells reveals deregulated expression of NKL homeobox genes HHEX and HLX in anaplastic large cell lymphoma (ALCL).Oncotarget. 2020 Aug 25;11(34):3208-3226. doi: 10.18632/oncotarget.27683. eCollection 2020 Aug 25. Oncotarget. 2020. PMID: 32922661 Free PMC article.
-
Type I interferons induce apoptosis by balancing cFLIP and caspase-8 independent of death ligands.Mol Cell Biol. 2013 Feb;33(4):800-14. doi: 10.1128/MCB.01430-12. Epub 2012 Dec 10. Mol Cell Biol. 2013. PMID: 23230268 Free PMC article.
-
Up-regulation of Smurf1 after spinal cord injury in adult rats.J Mol Histol. 2013 Aug;44(4):381-90. doi: 10.1007/s10735-013-9499-2. Epub 2013 Apr 18. J Mol Histol. 2013. PMID: 23595775
-
Molecular targets of TRAIL-sensitizing agents in colorectal cancer.Int J Mol Sci. 2012;13(7):7886-7901. doi: 10.3390/ijms13077886. Epub 2012 Jun 25. Int J Mol Sci. 2012. PMID: 22942679 Free PMC article. Review.
References
-
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP (2009) Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 9: 862–873 - PubMed
-
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501 - PubMed
-
- Burns TF, Bernhard EJ, El-Deiry WS (2001) Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 20: 4601–4612 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

