Evidence for dynamics in proteins as a mechanism for ligand dissociation
- PMID: 22246400
- PMCID: PMC3288659
- DOI: 10.1038/nchembio.769
Evidence for dynamics in proteins as a mechanism for ligand dissociation
Abstract
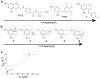
Signal transduction, regulatory processes and pharmaceutical responses are highly dependent upon ligand residence times. Gaining insight into how physical factors influence residence times (1/k(off)) should enhance our ability to manipulate biological interactions. We report experiments that yield structural insight into k(off) involving a series of eight 2,4-diaminopyrimidine inhibitors of dihydrofolate reductase whose binding affinities vary by six orders of magnitude. NMR relaxation-dispersion experiments revealed a common set of residues near the binding site that undergo a concerted millisecond-timescale switching event to a previously unidentified conformation. The rate of switching from ground to excited conformations correlates exponentially with the binding affinity K(i) and k(off), suggesting that protein dynamics serves as a mechanical initiator of ligand dissociation within this series and potentially for other macromolecule-ligand systems. Although the forward rate of conformational exchange, k(conf,forward), is faster than k(off), the use of the ligand series allowed for connections to be drawn between kinetic events on different timescales.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
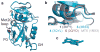


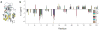
References
-
- Tummino PJ, Copeland RA. Residence time of receptor-ligand complexes and its effect on biological function. Biochemistry. 2008;47:5481–5492. - PubMed
-
- Olson JS, Phillips GN., Jr Kinetic pathways and barriers for ligand binding to myoglobin. J Biol Chem. 1996;271:17593–17596. - PubMed
-
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

