Chemical chaperones assist intracellular folding to buffer mutational variations
- PMID: 22246401
- PMCID: PMC3527004
- DOI: 10.1038/nchembio.768
Chemical chaperones assist intracellular folding to buffer mutational variations
Abstract
Hidden genetic variations have the potential to lead to the evolution of new traits. Molecular chaperones, which assist protein folding, may conceal genetic variations in protein-coding regions. Here we investigate whether the chemical milieu of cells has the potential to alleviate intracellular protein folding, a possibility that could implicate osmolytes in concealing genetic variations. We found that the model osmolyte trimethylamine N-oxide (TMAO) can buffer mutations that impose kinetic traps in the folding pathways of two model proteins. Using this information, we rationally designed TMAO-dependent mutants in vivo, starting from a TMAO-independent protein. We show that different osmolytes buffer a unique spectrum of mutations. Consequently, the chemical milieu of cells may alter the folding pathways of unique mutant variants in polymorphic populations and lead to unanticipated spectra of genetic buffering.
Figures
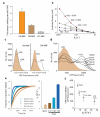
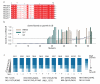



Comment in
-
Protein folding: Chaperoning protein evolution.Nat Chem Biol. 2012 Feb 15;8(3):226-8. doi: 10.1038/nchembio.791. Nat Chem Biol. 2012. PMID: 22337093 No abstract available.
References
-
- DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–87. - PubMed
-
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. - PubMed
-
- Waddington C. Canalization of Development and the Inheritance of Acquired Characters. Nature. 1942;150:563–565.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

