Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site
- PMID: 22247292
- PMCID: PMC3306683
- DOI: 10.1073/pnas.1111303109
Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site
Abstract
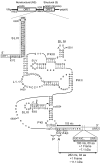
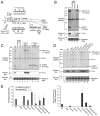
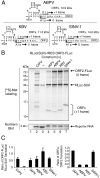
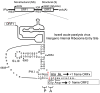
The dicistrovirus intergenic region internal ribosome entry site (IRES) utilizes a unique mechanism, involving P-site tRNA mimicry, to directly assemble 80S ribosomes and initiate translation at a specific non-AUG codon in the ribosomal A site. A subgroup of dicistrovirus genomes contains an additional stem-loop 5'-adjacent to the IRES and a short open reading frame (ORFx) that overlaps the viral structural polyprotein ORF (ORF2) in the +1 reading frame. Using mass spectrometry and extensive mutagenesis, we show that, besides directing ORF2 translation, the Israeli acute paralysis dicistrovirus IRES also directs ORFx translation. The latter is mediated by a UG base pair adjacent to the P-site tRNA-mimicking domain. An ORFx peptide was detected in virus-infected honey bees by multiple reaction monitoring mass spectrometry. Finally, the 5' stem-loop increases IRES activity and may couple translation of the two major ORFs of the virus. This study reveals a novel viral strategy in which a tRNA-like IRES directs precise, initiator Met-tRNA-independent translation of two overlapping ORFs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bonning BC, Miller WA. Dicistroviruses. Annu Rev Entomol. 2010;55:129–150. - PubMed
-
- Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. - PubMed
-
- Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

