Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins
- PMID: 22247555
- PMCID: PMC3293521
- DOI: 10.1074/jbc.M111.322958
Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins
Abstract
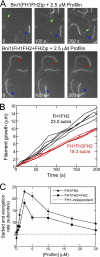
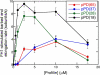
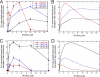
Formin-mediated elongation of actin filaments proceeds via association of Formin Homology 2 (FH2) domain dimers with the barbed end of the filament, allowing subunit addition while remaining processively attached to the end. The flexible Formin Homology 1 (FH1) domain, located directly N-terminal to the FH2 domain, contains one or more stretches of polyproline that bind the actin-binding protein profilin. Diffusion of FH1 domains brings associated profilin-actin complexes into contact with the FH2-bound barbed end of the filament, thereby enabling direct transfer of actin. We investigated how the organization of the FH1 domain of budding yeast formin Bni1p determines the rates of profilin-actin transfer onto the end of the filament. Each FH1 domain transfers actin to the barbed end independently of the other and structural evidence suggests a preference for actin delivery from each FH1 domain to the closest long-pitch helix of the filament. The transfer reaction is diffusion-limited and influenced by the affinities of the FH1 polyproline tracks for profilin. Position-specific sequence variations optimize the efficiency of FH1-stimulated polymerization by binding profilin weakly near the FH2 domain and binding profilin more strongly farther away. FH1 domains of many other formins follow this organizational trend. This particular sequence architecture may optimize the efficiency of FH1-stimulated elongation.
Figures


References
-
- Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. (1999) Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143 - PubMed
-
- Pellegrin S., Mellor H. (2005) The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol.: CB 15, 129–133 - PubMed
-
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., Boone C. (2002) Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

