The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay
- PMID: 22248237
- PMCID: PMC3273433
- DOI: 10.1186/1471-2199-13-1
The double-stranded break-forming activity of plant SPO11s and a novel rice SPO11 revealed by a Drosophila bioassay
Abstract
Background: SPO11 is a key protein for promoting meiotic recombination, by generating chromatin locus- and timing-specific DNA double-strand breaks (DSBs). The DSB activity of SPO11 was shown by genetic analyses, but whether SPO11 exerts DSB-forming activity by itself is still an unanswered question. DSB formation by SPO11 has not been detected by biochemical means, probably because of a lack of proper protein-folding, posttranslational modifications, and/or specific SPO11-interacting proteins required for this activity. In addition, plants have multiple SPO11-homologues.
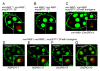
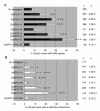
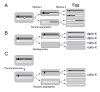
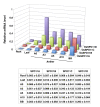
Results: To determine whether SPO11 can cleave DNA by itself, and to identify which plant SPO11 homologue cleaves DNA, we developed a Drosophila bioassay system that detects the DSB signals generated by a plant SPO11 homologue expressed ectopically. We cytologically and genetically demonstrated the DSB activities of Arabidopsis AtSPO11-1 and AtSPO11-2, which are required for meiosis, in the absence of other plant proteins. Using this bioassay, we further found that a novel SPO11-homologue, OsSPO11D, which has no counterpart in Arabidopsis, displays prominent DSB-forming activity. Quantitative analyses of the rice SPO11 transcripts revealed the specific increase in OsSPO11D mRNA in the anthers containing meiotic pollen mother cells.
Conclusions: The Drosophila bioassay system successfully demonstrated that some plant SPO11 orthologues have intrinsic DSB activities. Furthermore, we identified a novel SPO11 homologue, OsSPO11D, with robust DSB activity and a possible meiotic function.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

