Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus
- PMID: 22249968
- PMCID: PMC3277630
- DOI: 10.1136/amiajnl-2011-000597
Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus
Abstract
Objective: To evaluate data fragmentation across healthcare centers with regard to the accuracy of a high-throughput clinical phenotyping (HTCP) algorithm developed to differentiate (1) patients with type 2 diabetes mellitus (T2DM) and (2) patients with no diabetes.
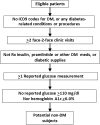
Materials and methods: This population-based study identified all Olmsted County, Minnesota residents in 2007. We used provider-linked electronic medical record data from the two healthcare centers that provide >95% of all care to County residents (ie, Olmsted Medical Center and Mayo Clinic in Rochester, Minnesota, USA). Subjects were limited to residents with one or more encounter January 1, 2006 through December 31, 2007 at both healthcare centers. DM-relevant data on diagnoses, laboratory results, and medication from both centers were obtained during this period. The algorithm was first executed using data from both centers (ie, the gold standard) and then from Mayo Clinic alone. Positive predictive values and false-negative rates were calculated, and the McNemar test was used to compare categorization when data from the Mayo Clinic alone were used with the gold standard. Age and sex were compared between true-positive and false-negative subjects with T2DM. Statistical significance was accepted as p<0.05.
Results: With data from both medical centers, 765 subjects with T2DM (4256 non-DM subjects) were identified. When single-center data were used, 252 T2DM subjects (1573 non-DM subjects) were missed; an additional false-positive 27 T2DM subjects (215 non-DM subjects) were identified. The positive predictive values and false-negative rates were 95.0% (513/540) and 32.9% (252/765), respectively, for T2DM subjects and 92.6% (2683/2898) and 37.0% (1573/4256), respectively, for non-DM subjects. Age and sex distribution differed between true-positive (mean age 62.1; 45% female) and false-negative (mean age 65.0; 56.0% female) T2DM subjects.
Conclusion: The findings show that application of an HTCP algorithm using data from a single medical center contributes to misclassification. These findings should be considered carefully by researchers when developing and executing HTCP algorithms.
Conflict of interest statement
Figures

References
-
- Delay No More: Improve Patient Recruitment and Reduce Time to Market in the Pharmaceutical Industry. https://www-935.ibm.com/services/sg/index.wss/ibvstudy/igs/x1014229?cntx...
-
- Wilke RA, Berg RL, Linneman JG, et al. Characterization of low-density lipoprotein cholesterol-lowering efficacy for atorvastatin in a population-based DNA biorepository. Basic Clin Pharmacol Toxicol 2008;103:354–9 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

