Postmortem cardiac tissue maintains gene expression profile even after late harvesting
- PMID: 22251372
- PMCID: PMC3342086
- DOI: 10.1186/1471-2164-13-26
Postmortem cardiac tissue maintains gene expression profile even after late harvesting
Abstract
Background: Gene expression studies can be used to help identify disease-associated genes by comparing the levels of expressed transcripts between cases and controls, and to identify functional genetic variants (expression quantitative loci or eQTLs) by comparing expression levels between individuals with different genotypes. While many of these studies are performed in blood or lymphoblastoid cell lines due to tissue accessibility, the relevance of expression differences in tissues that are not the primary site of disease is unclear. Further, many eQTLs are tissue specific. Thus, there is a clear and compelling need to conduct gene expression studies in tissues that are specifically relevant to the disease of interest. One major technical concern about using autopsy-derived tissue is how representative it is of physiologic conditions, given the effect of postmortem interval on tissue degradation.
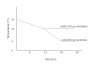
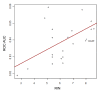
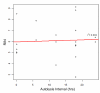
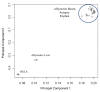
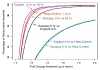
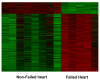
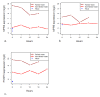
Results: In this study, we monitored the gene expression of 13 tissue samples harvested from a rapid autopsy heart (non-failed heart) and 7 from a cardiac explant (failed heart) through 24 hours of autolysis. The 24 hour autopsy simulation was designed to reflect a typical autopsy scenario where a body may begin cooling to ambient temperature for ~12 hours, before transportation and storage in a refrigerated room in a morgue. In addition, we also simulated a scenario wherein the body was left at room temperature for up to 24 hours before being found. A small fraction (< 2.5%) of genes showed fluctuations in expression over the 24 hr period and largely belong to immune and signal response and energy metabolism-related processes. Global expression analysis suggests that RNA expression is reproducible over 24 hours of autolysis with 95% genes showing < 1.2 fold change. Comparing the rapid autopsy to the failed heart identified 480 differentially expressed genes, including several types of collagens, lumican (LUM), natriuretic peptide A (NPPA) and connective tissue growth factor (CTGF), which allows for the clear separation between failing and non-failing heart based on gene expression profiles.
Conclusions: Our results demonstrate that RNA from autopsy-derived tissue, even up to 24 hours of autolysis, can be used to identify biologically relevant expression pattern differences, thus serving as a practical source for gene expression experiments.
Figures

Similar articles
-
Targeted myocardial gene expression in failing hearts by RNA sequencing.J Transl Med. 2016 Nov 25;14(1):327. doi: 10.1186/s12967-016-1083-6. J Transl Med. 2016. PMID: 27884156 Free PMC article.
-
Characterization of gene expression profiling of mouse tissues obtained during the postmortem interval.Exp Mol Pathol. 2016 Jun;100(3):482-92. doi: 10.1016/j.yexmp.2016.05.007. Epub 2016 May 13. Exp Mol Pathol. 2016. PMID: 27185020
-
Metabolic gene expression in fetal and failing human heart.Circulation. 2001 Dec 11;104(24):2923-31. doi: 10.1161/hc4901.100526. Circulation. 2001. PMID: 11739307
-
Life and death: A systematic comparison of antemortem and postmortem gene expression.Gene. 2020 Mar 20;731:144349. doi: 10.1016/j.gene.2020.144349. Epub 2020 Jan 11. Gene. 2020. PMID: 31935499
-
Global gene expression profiling in the failing myocardium.Circ J. 2009 Sep;73(9):1568-76. doi: 10.1253/circj.cj-09-0465. Epub 2009 Jul 29. Circ J. 2009. PMID: 19638707 Review.
Cited by
-
Complex Sources of Variation in Tissue Expression Data: Analysis of the GTEx Lung Transcriptome.Am J Hum Genet. 2016 Sep 1;99(3):624-635. doi: 10.1016/j.ajhg.2016.07.007. Am J Hum Genet. 2016. PMID: 27588449 Free PMC article.
-
Extensive Changes in Transcriptomic "Fingerprints" and Immunological Cells in the Large Organs of Patients Dying of Acute Septic Shock and Multiple Organ Failure Caused by Neisseria meningitidis.Front Cell Infect Microbiol. 2020 Feb 19;10:42. doi: 10.3389/fcimb.2020.00042. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32154187 Free PMC article.
-
Molecular Characterization of Pediatric Restrictive Cardiomyopathy from Integrative Genomics.Sci Rep. 2017 Jan 18;7:39276. doi: 10.1038/srep39276. Sci Rep. 2017. PMID: 28098235 Free PMC article.
-
Altered gene expression and possible immunodeficiency in cases of sudden infant death syndrome.Pediatr Res. 2016 Jul;80(1):77-84. doi: 10.1038/pr.2016.45. Epub 2016 Mar 9. Pediatr Res. 2016. PMID: 26959483
-
Rapid autopsies to enhance metastatic research: the UPTIDER post-mortem tissue donation program.NPJ Breast Cancer. 2024 Apr 24;10(1):31. doi: 10.1038/s41523-024-00637-3. NPJ Breast Cancer. 2024. PMID: 38658604 Free PMC article.
References
-
- Li JZ, Vawter MP, Walsh DM, Tomita H, Evans SJ, Choudary PV, Lopez JF, Avelar A, Shokoohi V, Chung T, Mesarwi O, Jones EG, Watson SJ, Akil H, Bunney WE Jr, Myers RM. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum Mol Genet. 2004;13:609–616. doi: 10.1093/hmg/ddh065. - DOI - PubMed
-
- Weinberger DR. Genetic mechanisms of psychosis: in vivo and postmortem genomics. Clin Ther. 2005;27(Suppl A):8–15. - PubMed
-
- Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, Overman KM, Atz ME, Myers RM, Jones EG, Watson SJ, Akil H, Bunney WE Jr. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

