Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML
- PMID: 22251480
- PMCID: PMC3311258
- DOI: 10.1182/blood-2011-10-386235
Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML
Abstract
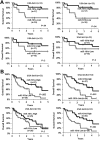
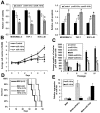
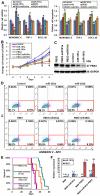
Increased expression levels of miR-181 family members have been shown to be associated with favorable outcome in patients with cytogenetically normal acute myeloid leukemia. Here we show that increased expression of miR-181a and miR-181b is also significantly (P < .05; Cox regression) associated with favorable overall survival in cytogenetically abnormal AML (CA-AML) patients. We further show that up-regulation of a gene signature composed of 4 potential miR-181 targets (including HOXA7, HOXA9, HOXA11, and PBX3), associated with down-regulation of miR-181 family members, is an independent predictor of adverse overall survival on multivariable testing in analysis of 183 CA-AML patients. The independent prognostic impact of this 4-homeobox-gene signature was confirmed in a validation set of 271 CA-AML patients. Furthermore, our in vitro and in vivo studies indicated that ectopic expression of miR-181b significantly promoted apoptosis and inhibited viability/proliferation of leukemic cells and delayed leukemogenesis; such effects could be reversed by forced expression of PBX3. Thus, the up-regulation of the 4 homeobox genes resulting from the down-regulation of miR-181 family members probably contribute to the poor prognosis of patients with nonfavorable CA-AML. Restoring expression of miR-181b and/or targeting the HOXA/PBX3 pathways may provide new strategies to improve survival substantially.
Figures

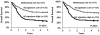
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

