IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome
- PMID: 22251702
- PMCID: PMC3266786
- DOI: 10.1172/JCI58847
IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome
Abstract
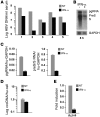
HBV infection remains a leading cause of death worldwide. IFN-α inhibits viral replication in vitro and in vivo, and pegylated IFN-α is a commonly administered treatment for individuals infected with HBV. The HBV genome contains a typical IFN-stimulated response element (ISRE), but the molecular mechanisms by which IFN-α suppresses HBV replication have not been established in relevant experimental systems. Here, we show that IFN-α inhibits HBV replication by decreasing the transcription of pregenomic RNA (pgRNA) and subgenomic RNA from the HBV covalently closed circular DNA (cccDNA) minichromosome, both in cultured cells in which HBV is replicating and in mice whose livers have been repopulated with human hepatocytes and infected with HBV. Administration of IFN-α resulted in cccDNA-bound histone hypoacetylation as well as active recruitment to the cccDNA of transcriptional corepressors. IFN-α treatment also reduced binding of the STAT1 and STAT2 transcription factors to active cccDNA. The inhibitory activity of IFN-α was linked to the IRSE, as IRSE-mutant HBV transcribed less pgRNA and could not be repressed by IFN-α treatment. Our results identify a molecular mechanism whereby IFN-α mediates epigenetic repression of HBV cccDNA transcriptional activity, which may assist in the development of novel effective therapeutics.
Figures
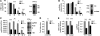
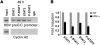



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

