Optimising multistage dairy cattle breeding schemes including genomic selection using decorrelated or optimum selection indices
- PMID: 22252172
- PMCID: PMC3292482
- DOI: 10.1186/1297-9686-44-1
Optimising multistage dairy cattle breeding schemes including genomic selection using decorrelated or optimum selection indices
Abstract
Background: The prediction of the outcomes from multistage breeding schemes is especially important for the introduction of genomic selection in dairy cattle. Decorrelated selection indices can be used for the optimisation of such breeding schemes. However, they decrease the accuracy of estimated breeding values and, therefore, the genetic gain to an unforeseeable extent and have not been applied to breeding schemes with different generation intervals and selection intensities in each selection path.
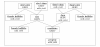
Methods: A grid search was applied in order to identify optimum breeding plans to maximise the genetic gain per year in a multistage, multipath dairy cattle breeding program. In this program, different values of the accuracy of estimated genomic breeding values and of their costs per individual were applied, whereby the total breeding costs were restricted. Both decorrelated indices and optimum selection indices were used together with fast multidimensional integration algorithms to produce results.
Results: In comparison to optimum indices, the genetic gain with decorrelated indices was up to 40% less and the proportion of individuals undergoing genomic selection was different. Additionally, the interaction between selection paths was counter-intuitive and difficult to interpret. Independent of using decorrelated or optimum selection indices, genomic selection replaced traditional progeny testing when maximising the genetic gain per year, as long as the accuracy of estimated genomic breeding values was ≥ 0.45. Overall breeding costs were mainly generated in the path "dam-sire". Selecting males was still the main source of genetic gain per year.
Conclusion: Decorrelated selection indices should not be used because of misleading results and the availability of accurate and fast algorithms for exact multidimensional integration. Genomic selection is the method of choice when maximising the genetic gain per year but genotyping females may not allow for a reduction in overall breeding costs. Furthermore, the economic justification of genotyping females remains questionable.
Figures


Similar articles
-
Optimum multistage genomic selection in dairy cattle.J Dairy Sci. 2012 Apr;95(4):2097-107. doi: 10.3168/jds.2011-4381. J Dairy Sci. 2012. PMID: 22459855
-
A comparison of dairy cattle breeding designs that use genomic selection.J Dairy Sci. 2011 Jan;94(1):493-500. doi: 10.3168/jds.2010-3518. J Dairy Sci. 2011. PMID: 21183061
-
Optimizing the design of small-sized nucleus breeding programs for dairy cattle with minimal performance recording.J Dairy Sci. 2014 Dec;97(12):7963-74. doi: 10.3168/jds.2014-8545. Epub 2014 Oct 3. J Dairy Sci. 2014. PMID: 25282422
-
Integrating genomic selection into dairy cattle breeding programmes: a review.Animal. 2013 May;7(5):705-13. doi: 10.1017/S1751731112002248. Epub 2012 Dec 3. Animal. 2013. PMID: 23200196 Review.
-
Genomic Selection in Dairy Cattle: The USDA Experience.Annu Rev Anim Biosci. 2017 Feb 8;5:309-327. doi: 10.1146/annurev-animal-021815-111422. Epub 2016 Nov 16. Annu Rev Anim Biosci. 2017. PMID: 27860491 Review.
Cited by
-
Community-based breeding programs can realize sustainable genetic gain and economic benefits in tropical dairy cattle systems.Front Genet. 2024 May 16;15:1106709. doi: 10.3389/fgene.2024.1106709. eCollection 2024. Front Genet. 2024. PMID: 38818034 Free PMC article.
-
Phenotypic and genetic analysis of milking temperament and its correlation with milk production traits in South African Holstein cattle.Trop Anim Health Prod. 2025 Apr 30;57(4):200. doi: 10.1007/s11250-025-04437-0. Trop Anim Health Prod. 2025. PMID: 40304851 Free PMC article.
-
The statistical theory of linear selection indices from phenotypic to genomic selection.Crop Sci. 2022 Mar-Apr;62(2):537-563. doi: 10.1002/csc2.20676. Epub 2022 Feb 6. Crop Sci. 2022. PMID: 35911794 Free PMC article.
-
Efficiency of a Constrained Linear Genomic Selection Index To Predict the Net Genetic Merit in Plants.G3 (Bethesda). 2019 Dec 3;9(12):3981-3994. doi: 10.1534/g3.119.400677. G3 (Bethesda). 2019. PMID: 31570501 Free PMC article.
-
Combined Multistage Linear Genomic Selection Indices To Predict the Net Genetic Merit in Plant Breeding.G3 (Bethesda). 2020 Jun 1;10(6):2087-2101. doi: 10.1534/g3.120.401171. G3 (Bethesda). 2020. PMID: 32312840 Free PMC article.
References
-
- Spelman R, Arias J, Keehan M, Obolonkin V, Winkelman A, Johnson D, Harris B. In: Proceedings of the 9th World Congress on Genetics Applied to Livestock Production: 1-6 August 2010; Leipzig. Gesellschaft fur Tierzuchtwissenschaften eV (German Society for Animal Science), editor. 2010. Application of genomic selection in the New Zealand dairy cattle industry. Abstract 0311.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

