Contribution of a TANK-binding kinase 1-interferon (IFN) regulatory factor 7 pathway to IFN-γ-induced gene expression
- PMID: 22252317
- PMCID: PMC3295005
- DOI: 10.1128/MCB.06021-11
Contribution of a TANK-binding kinase 1-interferon (IFN) regulatory factor 7 pathway to IFN-γ-induced gene expression
Abstract
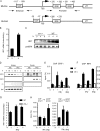
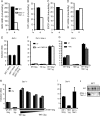
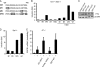
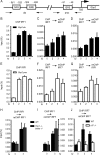
Signal transducers and activators of transcription (STATs) and interferon regulatory factors (IRFs) share common target genes. Here we show that the Irf7 gene is regulated by transcription factors STAT1 and IRF9 in response to the type II interferon (IFN) IFN-γ. IRF7 cooperated with STAT1 and IRF1 to stimulate the expression of a subset of IFN-γ-induced STAT1 target genes. IRF7-mediated control of the Gbp2 gene required the presence and basal activity of the S/T kinase TANK-binding kinase 1 (TBK1), whereas the binding of IRF7 to the Gbp2 promoter did not. Analysis of RNA polymerase II (Pol II) recruitment to the Gbp2 promoter revealed a role for IRF7 at later stages of the IFN-γ response. In support of the role of IRF7 in establishing an effective antibacterial response, IFN-γ-pretreated Irf7(-/-) macrophages showed an increased bacterial burden after infection with Listeria monocytogenes. Our data thus describe a biologically relevant basal activity of TBK1 and identify IRF7 as a novel player in the IFN-γ response.
Figures


Similar articles
-
Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-gamma-inducible gbp2 gene.Proc Natl Acad Sci U S A. 2007 Feb 20;104(8):2849-54. doi: 10.1073/pnas.0610944104. Epub 2007 Feb 9. Proc Natl Acad Sci U S A. 2007. PMID: 17293456 Free PMC article.
-
The C-Terminal Transactivation Domain of STAT1 Has a Gene-Specific Role in Transactivation and Cofactor Recruitment.Front Immunol. 2018 Dec 6;9:2879. doi: 10.3389/fimmu.2018.02879. eCollection 2018. Front Immunol. 2018. PMID: 30574148 Free PMC article.
-
Regulation of the transcriptional activity of the IRF7 promoter by a pathway independent of interferon signaling.J Biol Chem. 2005 Apr 1;280(13):12262-70. doi: 10.1074/jbc.M404260200. Epub 2005 Jan 21. J Biol Chem. 2005. PMID: 15664995
-
A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
-
Interferon regulatory factor 7 in inflammation, cancer and infection.Front Immunol. 2023 May 12;14:1190841. doi: 10.3389/fimmu.2023.1190841. eCollection 2023. Front Immunol. 2023. PMID: 37251373 Free PMC article. Review.
Cited by
-
Profiling of gallbladder carcinoma reveals distinct miRNA profiles and activation of STAT1 by the tumor suppressive miRNA-145-5p.Sci Rep. 2019 Mar 18;9(1):4796. doi: 10.1038/s41598-019-40857-3. Sci Rep. 2019. PMID: 30886199 Free PMC article.
-
TLR3 drives IRF6-dependent IL-23p19 expression and p19/EBI3 heterodimer formation in keratinocytes.Immunol Cell Biol. 2015 Oct;93(9):771-9. doi: 10.1038/icb.2015.77. Epub 2015 Aug 25. Immunol Cell Biol. 2015. PMID: 26303210
-
DNA methylation profiling identifies TBKBP1 as potent amplifier of cytotoxic activity in CMV-specific human CD8+ T cells.PLoS Pathog. 2024 Sep 26;20(9):e1012581. doi: 10.1371/journal.ppat.1012581. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39325839 Free PMC article.
-
RGS2 is an innate immune checkpoint for suppressing Gαq-mediated IFNγ generation and lung injury.iScience. 2025 Jan 27;28(2):111878. doi: 10.1016/j.isci.2025.111878. eCollection 2025 Feb 21. iScience. 2025. PMID: 40041768 Free PMC article.
-
Interferon gamma signaling positively regulates hematopoietic stem cell emergence.Dev Cell. 2014 Dec 8;31(5):640-53. doi: 10.1016/j.devcel.2014.11.007. Dev Cell. 2014. PMID: 25490269 Free PMC article.
References
-
- Au WC, Pitha PM. 2001. Recruitment of multiple interferon regulatory factors and histone acetyltransferase to the transcriptionally active interferon A promoters. J. Biol. Chem. 276:41629–41637 - PubMed
-
- Au WC, Yeow WS, Pitha PM. 2001. Analysis of functional domains of interferon regulatory factor 7 and its association with IRF-3. Virology 280:273–282 - PubMed
-
- Baccarini M, Bistoni F, Lohmann-Matthes ML. 1985. In vitro natural cell-mediated cytotoxicity against Candida albicans: macrophage precursors as effector cells. J. Immunol. 134:2658–2665 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
