snRNA 3' end formation requires heterodimeric association of integrator subunits
- PMID: 22252320
- PMCID: PMC3295014
- DOI: 10.1128/MCB.06511-11
snRNA 3' end formation requires heterodimeric association of integrator subunits
Abstract
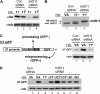
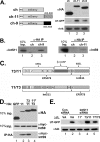
The Integrator Complex is a group of proteins responsible for the endonucleolytic cleavage of primary small nuclear RNA (snRNA) transcripts within the nucleus. Integrator subunits 9 and 11 (IntS9/11) are thought to contain the catalytic activity based on their high sequence similarity to CPSF100 and CPSF73, which have been shown to be components of both the poly(A)(+) and histone pre-mRNA cleavage complex. Here we demonstrate that the specific heterodimeric interaction between IntS9 and IntS11 is mediated by a discrete domain present at the extreme C terminus of IntS9 and within the C terminus of IntS11, adjacent to the predicted active site of this endonuclease. This domain is highly conserved within IntS11 but conspicuously absent in CPSF73. Using a cell-based complementation assay that measures Integrator activity, we determined that the IntS9 interaction domain within IntS11 is required for its ability to restore snRNA 3' end processing after RNA interference (RNAi)-mediated depletion of IntS11. Moreover, overexpression of these interaction domains alone elicits snRNA misprocessing through a dominant-negative titration of endogenous Integrator subunits. These data collectively explain the mechanism by which the IntS11/9 and, by analogy, the CPSF73/100 heterodimeric cleavage factors distinguish themselves from each other and demonstrate that the heterodimeric interaction is functionally required for snRNA 3' end formation.
Figures





References
-
- Aravind L. 1999. An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol. 1:69–91 - PubMed
-
- Baillat D, et al. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123:265–276 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
