Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7
- PMID: 22252322
- PMCID: PMC3295002
- DOI: 10.1128/MCB.06328-11
Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7
Abstract
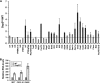
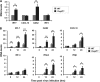
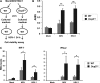
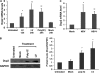
The mammalian Dcp2 mRNA-decapping protein functions primarily on a subset of mRNAs in a transcript-specific manner. Here we show that Dcp2 is an important modulator of genes involved in the type I interferon (IFN) response, which is the initial line of antiviral innate immune response elicited by a viral challenge. Mouse embryonic fibroblast cells with reduced Dcp2 levels (Dcp2(β/β)) contained significantly elevated levels of mRNAs encoding proteins involved in the type I IFN response. In particular, analysis of a key type I IFN transcription factor, IFN regulatory factor 7 (IRF-7), revealed an increase in both IRF-7 mRNA and protein in Dcp2(β/β) cells. Importantly, the increase in IRF-7 mRNA within the background of reduced Dcp2 levels was attributed to a stabilization of the IRF-7 mRNA, suggesting that Dcp2 normally modulates IRF-7 mRNA stability. Moreover, Dcp2 expression was also induced upon viral infection, consistent with a role in attenuating the antiviral response by promoting IRF-7 mRNA degradation. The induction of Dcp2 levels following a viral challenge and the specificity of Dcp2 in targeting the decay of IRF-7 mRNA suggest that Dcp2 may negatively contribute to the innate immune response in a negative feedback mechanism to restore normal homeostasis following viral infection.
Figures
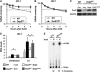
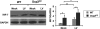
References
-
- Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. - PubMed
-
- Biggioggero M, Gabbriellini L, Meroni PL. 2010. Type I interferon therapy and its role in autoimmunity. Autoimmunity 43:248–254 - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891–1899 - PubMed
-
- Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
