ELL facilitates RNA polymerase II pause site entry and release
- PMID: 22252557
- PMCID: PMC3272570
- DOI: 10.1038/ncomms1652
ELL facilitates RNA polymerase II pause site entry and release
Abstract
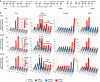
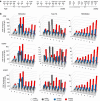
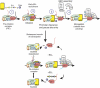
Transcription is a multi-stage process that coordinates several steps within the transcription cycle including chromatin reorganization, RNA polymerase II recruitment, initiation, promoter clearance and elongation. Recent advances have identified the super elongation complex, containing the eleven-nineteen lysine-rich leukaemia (ELL) protein, as a key regulator of transcriptional elongation. Here we show that ELL has a diverse and kinetically distinct role before its assembly into the super elongation complex by stabilizing Pol II recruitment/initiation and entry into the pause site. Loss of ELL destabilizes the pre-initiation complexes and results in disruption of early elongation and promoter proximal chromatin structure before recruitment of AFF4 and other super elongation complex components. These changes result in significantly reduced transcriptional activation of rapidly induced genes. Thus, ELL has an early and essential role during rapid high-amplitude gene expression that is required for both Pol II pause site entry and release.
Figures
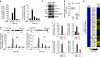




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

